Rapid Transmission and Divergence of Vancomycin-Resistant Enterococcus faecium Sequence Type 80, China
Liqiang Li
1, Xingwei Wang
1, Yanyu Xiao, Bing Fan, Jiehong Wei, Jie Zhou, Zetian Lai, Yanpeng Zhang, Hongmei Mo, Li Zhang, Dixian Luo, Dayong Gu, Shucai Yang, Yidi Wang, Jiuxin Qu

, and
Shenzhen Pathogen Infection Research Alliance (SPIRA)
Author affiliation: Shenzhen Third People’s Hospital Department of Clinical Laboratory; National Clinical Research Center for Infectious Diseases, Second Affiliated Hospital of Southern University of Science and Technology, Shenzhen, China (L. Li, X. Wang, Y. Xiao, Y. Wang, J. Qu); Shenzhen Institute of Translational Medicine, First Affiliated Hospital of Shenzhen University, Shenzhen Second People’s Hospital, Shenzhen (B. Fan, Y. Zhang, D. Gu); Medical Laboratory of the Third Affiliated Hospital of Shenzhen University, Shenzhen (J. Wei, H. Mo); Pingshan Hospital, Southern Medical University, Pingshan District People’s Hospital of Shenzhen, Shenzhen (J. Zhou, L. Zhang, S. Yang); Shenzhen Nanshan People's Hospital, Shenzhen (Z. Lai, D. Luo); Shenzhen University Medical School, Shenzhen (Z. Lai, D. Luo)
Main Article
Figure 1
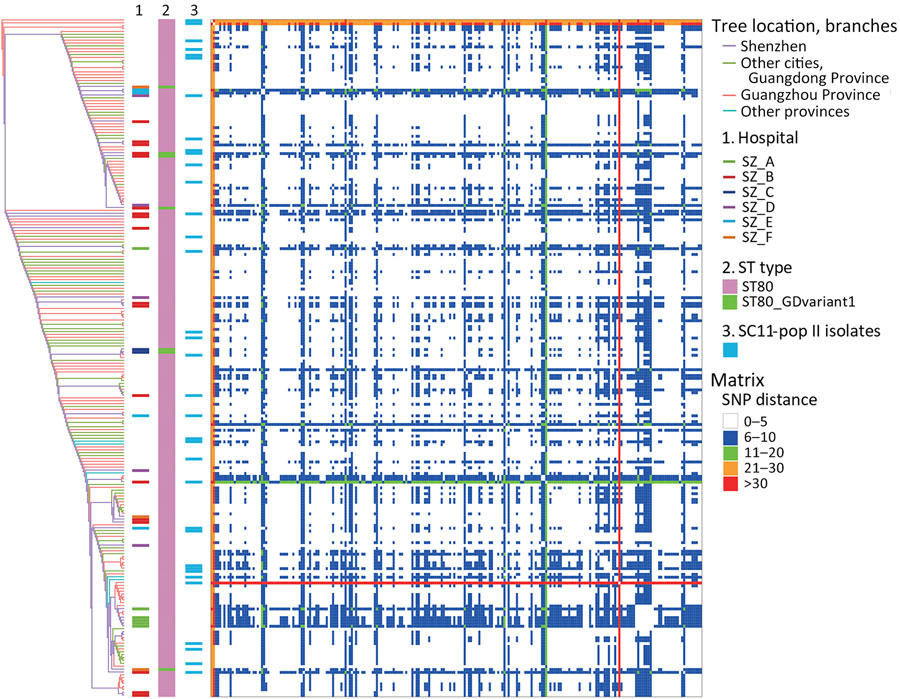
Figure 1. Evolution and variation of SC11 inferred from core genome SNPs during rapid transmission and divergence of vancomycin-resistant Enterococcus faecium ST80, China. Graphs shows reconstructed tree from core genome SNPs (left) among all SC11 isolates (n = 235) using strain SZYSC_GYSVRE003 (GenBank accession no. GCA_037475005.1) as reference. Hospital sources of isolates from Shenzhen ST80 and its variant, and SC11-pop II isolates are indicated (1, 2, and 3) next to the tree. The heatmap (right) shows pairwise SNP distance matrix indicating diversity of SC11 lineage presented in the form of symmetry in the bottom left and top right. Cells in the heatmap are colored to show SNP distance in a graded gradient. Red lines indicate large SNP distances and correspond to SZYSC_23VRE019 in the tree. pop, population; SC, sequence cluster; SNP, single-nucleotide polymorphism; ST, sequence type.
Main Article
Page created: March 24, 2025
Page updated: April 25, 2025
Page reviewed: April 25, 2025
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
