Volume 31, Number 7—July 2025
Research Letter
Next-Generation Sequencing Techniques to Diagnose Culture-Negative Subacute Native Aortic Endocarditis
Cite This Article
Citation for Media
Abstract
Next-generation sequencing might improve diagnosis of infective endocarditis. A case in Switzerland was initially attributed to Solobacterium moorei bacteria. Metagenomic analysis of the affected heart valve detected Streptococcus gordonii, but not S. moorei, illustrating that the results of molecular detection can vary depending on sampling time and anatomic site.
Plasma microbial cell-free DNA (mcfDNA) refers to extracellular microbial DNA in plasma, which has a half-life of a few minutes (1). Next-generation sequencing using mcfDNA is emerging as a diagnostic tool in infections with negative cultures, including endocarditis. Persistence of mcfDNA is associated with metastatic infection (2). We used next-generation mcfDNA sequencing to identify the causative agent in a fatal case of infective endocarditis.
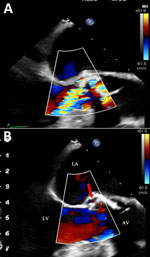
An 89-year-old man with aortic stenosis and preserved heart function sought care for weakness in Geneva, Switzerland. He had recently sought care at University Hospital of the Canary Islands (Tenerife, Spain) after a fall; elevated troponin (2,172 ng/L) and procalcitonin (157 mg/L) were observed. He received empiric meropenem and linezolid before he returned to Switzerland against medical advice. In Geneva, he had no fever or peripheral signs of endocarditis. Investigations revealed mild inflammation (C-reactive protein 17 mg/L), acute kidney injury (creatinine 387 µmol/L), stroke, and carotid stenosis. Transesophageal echocardiography showed an aortic valve perforation and a 4 mm para-aortic abscess at the root of the aorta, without suspected vegetation (Figure 1). Blood cultures remained negative after 5 days. We started conservative treatment with ceftriaxone (2 g every 12 h intravenously) and vancomycin (15 mg/kg every 12 h intravenously). mcfDNA next-generation sequencing (Noscendo, https://noscendo.com) identified Solobacterium moorei (13 reads). We adjusted the patient’s treatment to ceftriaxone (2 g every 12 h intravenously) and metronidazole (500 mg every 6 h intravenously) for 6 weeks.
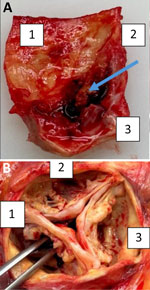
Despite initial improvement, the patient experienced heart failure and a second-degree AV block. His condition declined 12 weeks later, and he died. At autopsy, the heart showed a heavily calcified, perforated noncoronary leaflet of the aortic valve with a 15 × 10-mm blood-filled neocavity beneath it, extending to the valvular ring (Figure 2). Although we did not detect pus, our findings strongly suggested infective endocarditis because degenerative processes do not typically cause valve perforation or cavity formation. Those conditions are consistent with infective endocarditis (IE). A second mcfDNA test detected no bacteria.
We performed metagenomic next-generation sequencing (mNGS) of the valve tissue as previously described (3). We identified 1.28 million human reads and 36,629 reads from the spiked (8.5 × 104) control organism Bacillus spizizenii, along with 4,654 reads from Streptococcus gordonii, 146 reads from S. sanguinis, and 11 reads from Cutibacterium acnes (European Nucleotide Archive accession no. PRJEB81450). We used MetaPhlAn2 (https://huttenhower.sph.harvard.edu/metaphlan2) to confirm S. gordonii, which suggested it was the dominant pathogen in tissue (4). Reads identified as S. sanguinis were likely S. gordonii as well because of their high genomic similarity. We detected no Solobacterium moorei in the tissue. S. moorei is a gram-positive anaerobic rod from oral and intestinal microbiota. Although rarely detected, it has been implicated in human infections, especially in immunocompromised patients (5–7). Its identification is difficult because of its slow growth. It is generally susceptible to antimicrobial drugs for anaerobic infections, although resistance to rifampin and moxifloxacin has been reported (8).
This case demonstrates the utility of mcfDNA and metagenomic sequencing in culture-negative endocarditis. After negative routine work-up, we performed mcfDNA because the conservative management prevented valve resection. Although S. moorei was detected in plasma initially, a follow-up mcfDNA test 6 weeks after antimicrobial treatment was negative. That result likely indicates bacterial clearance, because it slightly exceeds the median 38-day positivity duration observed in infective endocarditis (9). S. gordonii was the only pathogen identified in valve tissue. The discrepancy between cfDNA and mNGS may reflect differing bacterial loads, sampling timing, or antimicrobial impact (10).
Our findings suggest an endocarditis caused by both S. gordonii and S. moorei organisms in which S. moorei mcfDNA predominated during the initial sampling but its culture likely failed because of antimicrobial exposure. In contrast, S. gordonii DNA seemed to persist longer in the valve tissue, suggesting greater stability in that environment. The absence of S. moorei in the valve tissue raises questions about its pathogenic role; its presence in mcfDNA could represent a transient bacteremia, another unrelated site of infection, or a contamination, but its relative abundance may have masked initial detection of S. gordonii bacteria. S. gordonii is a known endocarditis pathogen causing destructive IE, and its pathogenic role is therefore highly probable.
In case of a high suspicion of IE and when surgery is not feasible, we advise collecting additional blood or plasma samples at least 2 times within the first 24–48 hours. If blood cultures yield negative results, stored samples can undergo mcfDNA analysis. Testing multiple samples improves diagnostic reliability by minimizing the risk for unrelated transient bacteremia or contamination. If valve removal occurs, mNGS should be done as a final test for pathogen identification.
In summary, we report a case of destructive native aortic valve endocarditis without fever or marked inflammation. mcfDNA and mNGS were essential to identify the pathogen. Molecular diagnostics are valuable in culture-negative infections, particularly when conventional methods and tissue sampling are limited.
Dr. Vetterli is a chief resident in the primary care medicine department at the University Hospitals of Geneva. She has a special interest in mentorship and teaching. Dr. Zennaro is a resident physician in the internal medicine department at the University Hospitals of Geneva.
References
- Grumaz S, Stevens P, Grumaz C, Decker SO, Weigand MA, Hofer S, et al. Next-generation sequencing diagnostics of bacteremia in septic patients. Genome Med. 2016;8:73. DOIPubMedGoogle Scholar
- Eichenberger EM, de Vries CR, Ruffin F, Sharma-Kuinkel B, Park L, Hong D, et al. Microbial cell-free DNA identifies etiology of bloodstream infections, persists longer than conventional blood cultures, and its duration of detection is associated with metastatic infection in patients with Staphylococcus aureus and gram-negative bacteremia. Clin Infect Dis. 2022;74:2020–7. DOIPubMedGoogle Scholar
- Ramakrishnan G, Kronig I, Gaïa N, Lazarevic V, Schrenzel J. Mycoplasma genitalium endocarditis in prosthetic aortic valve. Emerg Infect Dis. 2023;29:2164–6. DOIPubMedGoogle Scholar
- Manghi P, Blanco-Míguez A, Manara S, NabiNejad A, Cumbo F, Beghini F, et al. MetaPhlAn 4 profiling of unknown species-level genome bins improves the characterization of diet-associated microbiome changes in mice. Cell Rep. 2023;42:
112464 . DOIPubMedGoogle Scholar - Liu WJ, Xiao M, Yi J, Li Y, Kudinha T, Xu YC. First case report of bacteremia caused by Solobacterium moorei in China, and literature review. BMC Infect Dis. 2019;19:730. DOIPubMedGoogle Scholar
- Alauzet C, Aujoulat F, Lozniewski A, Ben Brahim S, Domenjod C, Enault C, et al. A new look at the genus Solobacterium: a retrospective analysis of twenty-seven cases of infection involving S. moorei and a review of sequence databases and the literature. Microorganisms. 2021;9:1229. DOIPubMedGoogle Scholar
- Detry G, Pierard D, Vandoorslaer K, Wauters G, Avesani V, Glupczynski Y. Septicemia due to Solobacterium moorei in a patient with multiple myeloma. Anaerobe. 2006;12:160–2. DOIPubMedGoogle Scholar
- Pedersen RM, Holt HM, Justesen US. Solobacterium moorei bacteremia: identification, antimicrobial susceptibility, and clinical characteristics. J Clin Microbiol. 2011;49:2766–8. DOIPubMedGoogle Scholar
- Lazarevic V, Gaïa N, Pham TT, de Lorenzi-Tognon M, Girard M, Mauffrey F, et al. Identification of causative agents of infective endocarditis by metagenomic next-generation sequencing of resected valves. Front Cell Infect Microbiol. 2025;15:
1532257 . DOIPubMedGoogle Scholar - Eichenberger EM, Degner N, Scott ER, Ruffin F, Franzone J, Sharma-Kuinkel B, et al. Microbial cell-free DNA identifies the causative pathogen in infective endocarditis and remains detectable longer than conventional blood culture in patients with prior antibiotic therapy. Clin Infect Dis. 2023;76:e1492–500. DOIPubMedGoogle Scholar
Figures
Cite This ArticleOriginal Publication Date: June 20, 2025
1These authors are co–first authors.
Table of Contents – Volume 31, Number 7—July 2025
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Jacques Schrenzel, Division of Infectious Diseases, Department of Medicine, Geneva University Hospitals and University of Geneva, Geneva, Switzerland
Top