Volume 31, Number 7—July 2025
Dispatch
Outbreak of Ceftriaxone-Resistant Salmonella enterica Serovar Typhi, Bangladesh, 2024
Figure 3
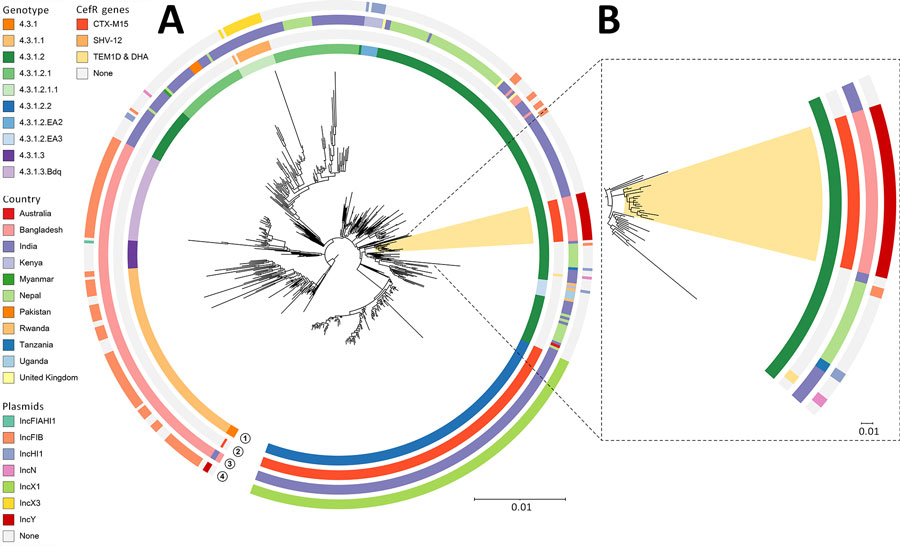
Figure 3. Phylogenetic tree of Salmonella Typhi genotype 4.3.1, including the 17 CefR 4.3.1.2.B1 strains detected in findings from a study of an outbreak of CefR S. enterica serovar Typhi, Bangladesh, 2024. A) Phylogenetic tree of 546 genomes belonging to genotype 4.3.1 and its subtypes. CefR strains sequenced in our study belong to genotype 4.3.1 and are highlighted in yellow. Tree was built following a pipeline described earlier (10) and displays different CefR genes, countries of isolation, and associated plasmid elements. For context, 529 genomes from genotype 4.3.1 and its subtypes were also included. Of those, 249 (10%) were randomly selected from 2,567 genomes (genotype 4.3.1.2 and subtypes) available on Pathogenwatch by genotype, year, and country (accessions available for 2,542; accessed on 14 July 2024), and 280 were from previous studies conducted in Bangladesh, India, and Pakistan (14). B) Zoomed-in view of the subclade containing the CefR strains from our study in Bangladesh. Scale bars indicate mean nucleotide substitutions per site. CefR, ceftriaxone-resistant.
References
- Stanaway JD, Reiner RC, Blacker BF, Goldberg EM, Khalil IA, Troeger CE, et al.; GBD 2017 Typhoid and Paratyphoid Collaborators. The global burden of typhoid and paratyphoid fevers: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Infect Dis. 2019;19:369–81. DOIPubMedGoogle Scholar
- Hooda Y, Tanmoy AM, Sajib MSI, Saha S. Mass azithromycin administration: considerations in an increasingly resistant world. BMJ Glob Health. 2020;5:
e002446 . DOIPubMedGoogle Scholar - Klemm EJ, Shakoor S, Page AJ, Qamar FN, Judge K, Saeed DK, et al. Emergence of an extensively drug-resistant Salmonella enterica serovar Typhi clone harboring a promiscuous plasmid encoding resistance to fluoroquinolones and third-generation cephalosporins. MBio. 2018;9:e00105–18. DOIPubMedGoogle Scholar
- Djeghout B, Saha S, Sajib MSI, Tanmoy AM, Islam M, Kay GL, et al. Ceftriaxone-resistant Salmonella Typhi carries an IncI1-ST31 plasmid encoding CTX-M-15. J Med Microbiol. 2018;67:620–7. DOIPubMedGoogle Scholar
- Argimón S, Nagaraj G, Shamanna V, Sravani D, Vasanth AK, Prasanna A, et al. Circulation of third-generation cephalosporin resistant Salmonella Typhi in Mumbai, India. Clin Infect Dis. 2022;74:2234–7. DOIPubMedGoogle Scholar
- Godbole G, McCann N, Jones SM, Dallman TJ, Brown M. Ceftriaxone-resistant Salmonella Typhi in a traveller returning from a mass gathering in Iraq. Lancet Infect Dis. 2019;19:467. DOIPubMedGoogle Scholar
- Saha S, Islam M, Uddin MJ, Saha S, Das RC, Baqui AH, et al. Integration of enteric fever surveillance into the WHO-coordinated Invasive Bacterial-Vaccine Preventable Diseases (IB-VPD) platform: A low cost approach to track an increasingly important disease. PLoS Negl Trop Dis. 2017;11:
e0005999 . DOIPubMedGoogle Scholar - Tanmoy AM, Hooda Y, Sajib MSI, Rahman H, Sarkar A, Das D, et al. Trends in antimicrobial resistance amongst Salmonella Typhi in Bangladesh: A 24-year retrospective observational study (1999-2022). PLoS Negl Trop Dis. 2024;18:
e0012558 . DOIPubMedGoogle Scholar - Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLOS Comput Biol. 2017;13:
e1005595 . DOIPubMedGoogle Scholar - Tanmoy AM, Hooda Y, Sajib MSI, da Silva KE, Iqbal J, Qamar FN, et al. Paratype: a genotyping tool for Salmonella Paratyphi A reveals its global genomic diversity. Nat Commun. 2022;13:7912. DOIPubMedGoogle Scholar
- Antipov D, Hartwick N, Shen M, Raiko M, Lapidus A, Pevzner PA. plasmidSPAdes: assembling plasmids from whole genome sequencing data. Bioinformatics. 2016;32:3380–7. DOIPubMedGoogle Scholar
- Wishart DS, Han S, Saha S, Oler E, Peters H, Grant JR, et al. PHASTEST: faster than PHASTER, better than PHAST. Nucleic Acids Res. 2023;51(W1):W443–50. DOIPubMedGoogle Scholar
- Carey ME, Dyson ZA, Ingle DJ, Amir A, Aworh MK, Chattaway MA, et al.; Global Typhoid Genomics Consortium Group Authorship. Global diversity and antimicrobial resistance of typhoid fever pathogens: Insights from a meta-analysis of 13,000 Salmonella Typhi genomes. eLife. 2023;12:
e85867 . DOIPubMedGoogle Scholar - Argimón S, David S, Underwood A, Abrudan M, Wheeler NE, Kekre M, et al.; NIHR Global Health Research Unit on Genomic Surveillance of Antimicrobial Resistance. Rapid genomic characterization and global surveillance of Klebsiella using Pathogenwatch. Clin Infect Dis. 2021;73(Suppl_4):S325–35. DOIPubMedGoogle Scholar