Volume 31, Number 7—July 2025
Dispatch
Outbreak of Ceftriaxone-Resistant Salmonella enterica Serovar Typhi, Bangladesh, 2024
Cite This Article
Citation for Media
Abstract
We report an outbreak of ceftriaxone-resistant Salmonella enterica serovar Typhi in Bangladesh; 47 cases were identified during April–September 2024. Isolates belonged to genotype 4.3.1.2 and harbored the blaCTX-M-15 gene on the pCROB1 plasmid. This genotype-plasmid lineage represents a recent introduction, calling for strengthened surveillance, antimicrobial stewardship, and vaccination strategies.
Salmonella enterica serovar Typhi, which causes typhoid fever, remains a major public health concern, particularly in South Asia, which accounts for ≈70% of global cases (1). Reports of drug-resistant Salmonella Typhi have increased in recent decades. Of particular concern are strains resistant to ceftriaxone, azithromycin, or both (2). In 2016, researchers identified an outbreak of extensively drug-resistant Salmonella Typhi in Pakistan (3), in which strains showed resistance to chloramphenicol, ampicillin, trimethoprim/sulfamethoxazole, fluoroquinolones, and third-generation cephalosporins (3). Additional reports noted sporadic cases of independently acquired ceftriaxone-resistant Salmonella Typhi from Bangladesh (4), India (5), and the United Kingdom (6). Routine use of ceftriaxone as empirical therapy in South Asia creates selective pressure, heightening the need for public health vigilance to prevent the spread of resistant Salmonella Typhi (2).
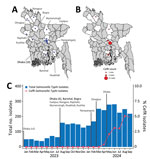
In Bangladesh, Salmonella Typhi is the most common cause of bloodstream infections in children >2 months of age; Dhaka, the capital city, is the primary focus of surveillance initiatives (7). To broaden the scope of surveillance and monitor the burden of disease and associated antibiotic drug susceptibility patterns nationwide, we began expanding our surveillance network in January 2023 to include clinics affiliated with Popular Diagnostic Centre Ltd (PDCL), a large, Bangladesh-based diagnostic services provider organization. As of May 2024, the network encompassed 20 clinics across 11 districts, including 10 in Dhaka (Figure 1, panel A). Leveraging this expanded passive surveillance for typhoid fever, we report data from January 2023–September 2024, describing the emergence, spread, and genomic epidemiology of a ceftriaxone-resistant clone of Salmonella Typhi in Bangladesh.
PDCL clinics are outpatient facilities that perform physician-prescribed blood cultures for patients. If a blood culture yields Salmonella Typhi, technicians transport the isolate to the Child Health Research Foundation (CHRF, https://chrfbd.org) laboratory for serovar confirmation and antibiotic drug susceptibility testing against ampicillin, cefixime, ceftazidime, ceftriaxone, meropenem, chloramphenicol, trimethoprim/sulfamethoxazole, ciprofloxacin, and azithromycin. Those processes follow established methodologies, including biochemical and slide-agglutination tests (Salmonella-agglutinating antiserum; Thermo Fisher Scientific, https://www. thermofisher.com) and Clinical and Laboratory Standards Institute–guided Kirby-Bauer disc diffusion methods (Oxoid; Thermo Fisher Scientific) (8).
We identified the first ceftriaxone-resistant Salmonella Typhi isolate on April 27, 2024, at the PDCL clinic in Narayanganj, a city 30 km from Dhaka, which was added to our surveillance network in February 2024. Subsequent monthly detection of ceftriaxone-resistant strains continued through the study period, culminating in 47 cases by September 2024. Of those, 41 cases were from Narayanganj and 6 were from other districts (Figure 1, panel B). The isolation rate of ceftriaxone-resistant Salmonella Typhi increased from 0% to 5% of all isolates during March 2024–September 2024 (Figure 1, panel C). All 47 isolates were resistant to amoxicillin and ceftriaxone, nonsusceptible to fluoroquinolones, but sensitive to chloramphenicol, trimethoprim/sulfamethoxazole, azithromycin, and meropenem (Table).
To investigate the basis of ceftriaxone resistance, we performed whole-genome sequencing on 17 of the 47 resistant Salmonella Typhi isolates (Appendix). Those isolates included 14 from Narayanganj and 1 each from Barishal, Mymensingh, and Rangpur. We prepared genomic libraries using the New England Biolabs DNA library preparation kit (New England Biolabs, https://www.neb.com) and sequenced on a NextSeq2000 platform (2 × 150 bp). We performed genome assembly using Unicycler v0.5.1 (9) and analyzed raw fastq files with Mykrobe v0.13.0 (D.J. Ingle et al., unpub. data, https://www.biorxiv.org/content/10.1101/2024.09.30.613582v1).
We classified all 17 ceftriaxone-resistant genomes as genotype 4.3.1.2, noting that ceftriaxone resistance was conferred by the blaCTX-M-15 gene, which showed 100% sequence identity with the gene found in extensively drug-resistant Salmonella Typhi strains from Pakistan (genotype 4.3.1.1.P1) (3). We performed phylogenetic analysis using Bowtie2, Samtools, Gubbins, and RAxML, following the pipeline described previously (10).
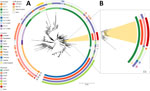
We identified an IncY plasmid in all 17 sequenced strains, which we further characterized using plasmidSPAdes v3.15.5 (11). We generated the longest plasmid sequence of 103.9 kbp from isolate STY_0313. This IncY plasmid, referred to as pCROB1, carried the blaCTX-M-15 gene and encoded a phage element (Figure 2). Public Health England previously identified a similar phage-plasmid associated with genotype 4.3.1.1 among travel-related typhoid cases from Iraq (6), suggesting a unique genotype-plasmid lineage underpinning the ongoing outbreak in Bangladesh.
Genotype 4.3.1.2 is rarely identified in Bangladesh and accounted for only 0.5% (7/1,356) of all Salmonella Typhi whole-genome sequences available for 1999–2018 (13). The genotype is more commonly found in India and Nepal (Figure 3). To investigate the phylogenetic context of genotype 4.3.1.2, we constructed a tree using global database sequences from genotype 4.3.1 and its subtypes, with a focus on 4.3.1.2 and its sublineages (Appendix). We found that the recent ceftriaxone resistant isolates from Bangladesh are closely related to strains from India and Nepal, rather than to earlier 4.3.1.2 genotypes from Bangladesh (Figure 3). However, our research showed no strains from India or Nepal were reported to be ceftriaxone-resistant or to carry the pCROB1 plasmid. We propose naming this distinct subclade lineage 4.3.1.2.B1. Although researchers have reported ceftriaxone resistance in genotype 4.3.1.2 strains from India (5), the molecular basis of resistance in lineage 4.3.1.2.B1 differs, suggesting an independent acquisition of the ceftriaxone-encoding phage-plasmid by this genotype.
To better understand the progression of disease in patients with ceftriaxone-resistant typhoid, we selected 35 cases (identified through August 31, 2024) for telephone interviews, successfully completing 21 (60%). Most (65%, 13/21) patients initially received either second-generation (cefuroxime 5%, 1/21) or third-generation (57%, 12/21; including cefixime [30%, 7/21], ceftriaxone [19%, 4/21], and ceftibuten [5%, 1/21]) cephalosporins. Forty-six percent (6/13) of patients switched to meropenem (23%, 3/13), azithromycin (8%, 1/13), or trimethoprim/sulfamethoxazole (8%, 1/13). One patient could not recall the second antibiotic used. All patients recovered; average illness duration was 19 (range 12–28) days. No patients reported travel outside Bangladesh, suggesting local circulation of this strain. Three cases occurred outside Narayanganj, the initial outbreak site; 2 patients had no travel history to Narayanganj within 15 days of illness onset, indicating potential spread to other districts.
Our detection of a unique genotype-plasmid lineage—4.3.1.2.B1, carrying the blaCTX-M-15 gene on the pCROB1 plasmid—associated with an outbreak of ceftriaxone-resistant Salmonella Typhi in Bangladesh represents a concerning development because of the strain’s potential for regional and international spread (2). Given the observed local spread of this lineage, healthcare systems may need to prepare for a shift back to older antibiotics, such as trimethoprim/sulfamethoxazole and chloramphenicol. Of note, 4.3.1.2.B1 strains remain sensitive to those drugs, indicating that first-line antibiotic drugs may serve as viable alternatives to azithromycin and meropenem. Seven patients in this study received third-generation cephalosporins and recovered from fever; however, we did not follow those cases to investigate relapse rates and other complications that could result from improper antibiotic use.
Our investigation of a unique strain of ceftriaxone-resistant Salmonella Typhi emerging in Bangladesh underscores the role of widespread ceftriaxone use in selecting for antimicrobial-resistant strains of typhoid fever. The outbreak was detected in April 2024, but the clone could have been circulating in Narayanganj before detection in our surveillance network. Because ceftriaxone remains a cornerstone of empirical treatment for typhoid, its declining efficacy is of great public health concern. Public health efforts should focus on bolstering antimicrobial stewardship and public education on appropriate antibiotic drug use, strengthening surveillance systems, and implementing and promoting immunization with typhoid conjugate vaccines to curb further spread of resistant strains.
Dr. Hooda is a senior scientist at the Child Health Research Foundation in Dhaka, Bangladesh, specializing in infectious diseases, antimicrobial resistance, and the development of innovative interventions like bacteriophages for public health challenges. His work focuses on advancing research capacity in low- and middle-income countries, including genomic analysis and pediatric infection monitoring, while fostering global collaborations to combat pressing health issues.
Acknowledgments
This article was preprinted at https://www.medrxiv.org/content/10.1101/2024.12.24.24319600v1.
We are grateful to the CHRF team, especially Md. Shariful Islam, Md. Shakiul Kabir, Lima Akter, Md. Zillur Rahman, Md. Humayun Kabir, Khursheda Afrin Khushi, and Monalisa, for their efforts in coordinating the timely transfer of Salmonella Typhi isolates and data from PDCL to CHRF, data management, aiding in the lab, and conducting phone follow-ups. This study would not have been possible without the invaluable support of the PDCL staff in facilitating the careful storage and transfer of the isolates.
Ethical approval for this study was obtained from the Ethics Review Boards of Bangladesh Shishu Hospital and Institute (IRB no. Admin/BSHI/2022/2058) and the CHRF (IRB no. CHRFIRB/07-12-2023/02). Raw fastq files of sequences from this study have been submitted to the European Nucleotide Archive (accession no. ERP167392).
This work was supported by the Bill & Melinda Gates Foundation (grant nos. INV-051975 and INV-008335).
References
- Stanaway JD, Reiner RC, Blacker BF, Goldberg EM, Khalil IA, Troeger CE, et al.; GBD 2017 Typhoid and Paratyphoid Collaborators. The global burden of typhoid and paratyphoid fevers: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Infect Dis. 2019;19:369–81. DOIPubMedGoogle Scholar
- Hooda Y, Tanmoy AM, Sajib MSI, Saha S. Mass azithromycin administration: considerations in an increasingly resistant world. BMJ Glob Health. 2020;5:
e002446 . DOIPubMedGoogle Scholar - Klemm EJ, Shakoor S, Page AJ, Qamar FN, Judge K, Saeed DK, et al. Emergence of an extensively drug-resistant Salmonella enterica serovar Typhi clone harboring a promiscuous plasmid encoding resistance to fluoroquinolones and third-generation cephalosporins. MBio. 2018;9:e00105–18. DOIPubMedGoogle Scholar
- Djeghout B, Saha S, Sajib MSI, Tanmoy AM, Islam M, Kay GL, et al. Ceftriaxone-resistant Salmonella Typhi carries an IncI1-ST31 plasmid encoding CTX-M-15. J Med Microbiol. 2018;67:620–7. DOIPubMedGoogle Scholar
- Argimón S, Nagaraj G, Shamanna V, Sravani D, Vasanth AK, Prasanna A, et al. Circulation of third-generation cephalosporin resistant Salmonella Typhi in Mumbai, India. Clin Infect Dis. 2022;74:2234–7. DOIPubMedGoogle Scholar
- Godbole G, McCann N, Jones SM, Dallman TJ, Brown M. Ceftriaxone-resistant Salmonella Typhi in a traveller returning from a mass gathering in Iraq. Lancet Infect Dis. 2019;19:467. DOIPubMedGoogle Scholar
- Saha S, Islam M, Uddin MJ, Saha S, Das RC, Baqui AH, et al. Integration of enteric fever surveillance into the WHO-coordinated Invasive Bacterial-Vaccine Preventable Diseases (IB-VPD) platform: A low cost approach to track an increasingly important disease. PLoS Negl Trop Dis. 2017;11:
e0005999 . DOIPubMedGoogle Scholar - Tanmoy AM, Hooda Y, Sajib MSI, Rahman H, Sarkar A, Das D, et al. Trends in antimicrobial resistance amongst Salmonella Typhi in Bangladesh: A 24-year retrospective observational study (1999-2022). PLoS Negl Trop Dis. 2024;18:
e0012558 . DOIPubMedGoogle Scholar - Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLOS Comput Biol. 2017;13:
e1005595 . DOIPubMedGoogle Scholar - Tanmoy AM, Hooda Y, Sajib MSI, da Silva KE, Iqbal J, Qamar FN, et al. Paratype: a genotyping tool for Salmonella Paratyphi A reveals its global genomic diversity. Nat Commun. 2022;13:7912. DOIPubMedGoogle Scholar
- Antipov D, Hartwick N, Shen M, Raiko M, Lapidus A, Pevzner PA. plasmidSPAdes: assembling plasmids from whole genome sequencing data. Bioinformatics. 2016;32:3380–7. DOIPubMedGoogle Scholar
- Wishart DS, Han S, Saha S, Oler E, Peters H, Grant JR, et al. PHASTEST: faster than PHASTER, better than PHAST. Nucleic Acids Res. 2023;51(W1):W443–50. DOIPubMedGoogle Scholar
- Carey ME, Dyson ZA, Ingle DJ, Amir A, Aworh MK, Chattaway MA, et al.; Global Typhoid Genomics Consortium Group Authorship. Global diversity and antimicrobial resistance of typhoid fever pathogens: Insights from a meta-analysis of 13,000 Salmonella Typhi genomes. eLife. 2023;12:
e85867 . DOIPubMedGoogle Scholar - Argimón S, David S, Underwood A, Abrudan M, Wheeler NE, Kekre M, et al.; NIHR Global Health Research Unit on Genomic Surveillance of Antimicrobial Resistance. Rapid genomic characterization and global surveillance of Klebsiella using Pathogenwatch. Clin Infect Dis. 2021;73(Suppl_4):S325–35. DOIPubMedGoogle Scholar
Figures
Table
Cite This ArticleOriginal Publication Date: June 20, 2025
Table of Contents – Volume 31, Number 7—July 2025
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Senjuti Saha, Child Health Research Foundation, SEL-Huq Skypark, 23/2 Khilji Rd, Block-B, Mohammadpur Dhaka, 1207, Bangladesh
Top