Volume 31, Number 8—August 2025
Research
Multidisciplinary Tracking of Highly Pathogenic Avian Influenza A(H5N1) Outbreak in Griffon Vultures, Southern Europe, 2022
Cite This Article
Citation for Media
Abstract
Since 2021, highly pathogenic avian influenza (HPAI) A(H5N1) clade 2.3.4.4b virus has affected wild bird populations globally. Griffon vultures (Gyps fulvus), a species hitherto considered unexposed, experienced an HPAI H5N1 outbreak in 2022 in southern Europe, leading to moderate mortality and reduced breeding success. The integration of virological, serologic, phylogenetic, and ecologic data revealed a short yet intense viral circulation and a probable common source of infection. The dissemination across Spain and France was likely caused by frequent interpopulation movements of birds. This integrated overview of the 2022 HPAI outbreak in vultures provides novel insights into the role of large-scale movements of wild birds in the spread of such disease. Understanding the epidemiologic dynamics of HPAI H5N1 in these scavenger species is crucial because the birds play vital roles in ecosystem functioning. Their susceptibility to this virus highlights potential broader ecologic effects of the ongoing outbreaks.
Since 2021, highly pathogenic avian influenza (HPAI) A(H5N1) clade 2.3.4.4b virus has emerged as a devastating pathogen in terms of bird species diversity, abundance, geographic extent, and economic losses (1). Although the effects on domestic birds have been staggering at >500 million reported deaths, the full extent of the toll on wild birds is unknown (2). Approximately 420,000 wild bird deaths have been reported, likely a considerable underestimate (3), and the diversity and number of affected species imply a profound threat to biodiversity (4).
This ongoing panzootic represents a paradigm shift in H5Nx avian influenza. HPAI H5N1 infections have now been reported on all continents except Oceania and in >50 mammal species (5,6). Most mammal infections have been reported in predators and scavengers, but livestock have also been affected, notably cattle in the United States (7). Furthermore, the loss of traditional seasonality, evidenced by outbreaks now persisting year-round, also represents a profound shift in HPAI virus ecology (8,9). Evidence for sustained mammal-to-mammal transmission is inconclusive, but the unprecedented geographic spread of the virus, coupled with the number of species infected, has raised concerns about the ability of the virus to expand its host range and gain pandemic potential (10).
Among wild birds, gregarious species, particularly those with colonial nesting behavior, have shown heightened vulnerability (11). Colonies of seabirds have experienced exceptionally high mortality rates worldwide (12–16). This susceptibility is likely because of enhanced virus transmission within densely aggregated avian populations, where the proximity of birds contributes to the rapid spread of the virus.
Vultures have been considered relatively resilient to pathogens, including HPAI virus, because of their scavenging diet and their associated physiologic adaptations to cope with pathogens (17). Before the current panzootic, only a local outbreak of HPAI had been reported in hooded vultures (Necrosyrtes monachus) in Burkina Faso in 2006 (18). Similarly, although ornitophagous raptors and scavengers have previously been only sporadically affected by HPAI H5Nx, they have been unexpectedly affected during the ongoing panzootic; birds affected have included obligate or occasional scavengers such as bald eagles (Haliaaetus leucocephalus), black vultures (Coragyps atratus), and California condors (Gymnogyps californianus) in North America and griffon vultures (Gyps fulvus) in Europe (19,20).
In spring 2022, abnormal deaths of nestling and adult griffon vultures were detected in Spain and France; HPAI H5N1 infection was confirmed by quantitative reverse transcription PCR (qRT-PCR) (Appendix Figure 1). This occurrence prompted an investigation into the epidemiology of HPAI in this vulture population. Leveraging ongoing ecologic studies, we investigated the origin of infection and assessed the nature and magnitude of viral spread through an integrated analysis of virological, serologic, genomic, and ecologic data obtained from field sampling in France and Spain. In addition to seroepidemiology and viral phylogenetic approaches, we further used long-term global positioning system (GPS) tracking data to evaluate potential sources of exposure, to study population connectivity pathways, and to investigate the potential of long-range contamination by movements of infected animals.
We collated and jointly analyzed information from griffon vultures collected from Spain and France in the framework of several research and surveillance programs dedicated to the study of vulture ecology, population dynamics, or HPAI outbreak follow-up (Appendix). In addition, the sampling included birds from national and local surveillance programs or those submitted for diagnosis to the institutions of the authors. In Spain, griffon vulture captures were performed in numerous colonies across the Iberian Peninsula during 2020–2022. Adult griffon vultures were captured with net traps or walk-in traps at vulture feeding stations, and nestlings were captured at nests. In France, 3 capture sessions were performed in 2022 and 2023 using walk-in traps at 5 sites.
All birds were ringed and a subset of vultures were fitted with GPS satellite transmitters. Blood samples, as well as oropharyngeal and cloacal swab samples, were collected from live birds. From dead birds, vascular feathers and tissues from the main organs (spleen, pancreas, heart, brain, trachea, intestine, lungs, and liver) were collected.
We extracted nucleic acids and submitted them to generic matrix gene qRT-PCR. We then submitted positive samples for nanopore sequencing (Oxford Nanopore Technologies, https://nanoporetech.com) and used consensus genomes in molecular marker and phylogeographic analysis.
We separated serum from the cell pellet by centrifugation in blood samples and stored at −20°C until analysis. We inactivated and tested serum samples with commercial ELISA and submitted positive samples to a hemagglutination inhibition (HI) test. We categorized HI samples as positive for an antigen if the titer was >16. We then estimated true seroprevalence by using the epidemiologic calculator Epitools (https://epitools.ausvet.com.au/prevalence).
We examined movement patterns of GPS-tagged griffon vultures during the 4-month period from March 1–June 29 during 2022 (outbreak year) and 2023 (control year), characterizing tracks as local or transit movements or immobility. We calculated daily distances traveled (DDT) and used generalized linear mixed models to identify factors affecting DDT. We first examined the movement patterns of a large sample of 114 birds, then examined the spatial and temporal dimensions of the movements of a subsample of 16 birds that remained in Europe during the study period.
HPAI H5N1 Outbreak Dynamics
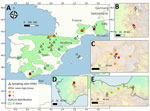
During April–August 2022, a total of 5 griffon vultures in Spain and 11 in France were confirmed to have died from HPAI H5N1 infection (Figure 1; Appendix). Despite the advanced state of decomposition, severe generalized congestion was detected on postmortem examination.
Apparently healthy live vultures sampled in both countries during March 2020–August 2023 tested negative for avian influenza virus (AIV) in oropharyngeal and cloacal swab samples (France, n = 393; Spain, n = 216). However, 2 birds admitted to a rehabilitation center in northern Spain (Figure 1, site 6) tested positive for HPAI H5 on May 5, 2022, and May 11, 2022. Both birds displayed weakness and severe central nervous system symptoms, including torticollis and inability to fly (Appendix). Only 1 of 87 live griffon vulture nestlings from Spain for which vascular feathers were available tested positive for HPAI H5 on May 24, 2022. The nestling, from southern Spain (site 14), lacked clinical signs at the time of ringing but was found dead on June 10, 2022; the remains tested positive for HPAI H5 (Appendix).
Exposure to H5 AIV in flying vultures was confirmed by the detection of H5-specific antibodies by HI tests after ELISA screening in 58 of 392 birds in France and 7 of 51 birds in Spain captured after the outbreak (summer 2022, autumn 2022, and summer 2023) (Table; Appendix Table 1). However, only 3 of 51 nestlings tested in Spain after the outbreak (summer 2022 and summer 2023) had H5-specific antibodies. Of 17 flying birds captured in Spain before the outbreak in spring 2020, a total of 8 were seropositive against strains of AIV other than H5 or H7, whereas none of the 50 nestlings tested before the outbreak (summer 2021) were seropositive against AIV. Of 128 H5 ELISA-positive vultures in France, 70 could not be confirmed by H5-specific HI tests.
Origin and Spatial Spread of Infection
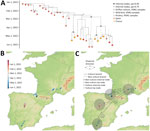
We performed phylogenetic analysis on a dataset of 12 hemagglutinin genetic sequences retrieved from HPAI H5N1–infected griffon vultures (8 generated in this study), together with a background dataset of sequences from poultry and wild birds that were sampled in Europe during November 8, 2021–September 1, 2022 (n = 571) (Appendix). The analysis confirmed that the sequences obtained in this study belong to clade 2.3.4.4b and showed that most vulture sequences were grouped into a distinct clade (n = 32) in a maximum-likelihood tree (bootstrap support = 0.83) (Appendix), with vulture sequences already identified as genotype AB (21). Mutation analysis of the vulture sequences showed that several variable sites were identified in the hemagglutinin segment, some of which have been previously associated with specific phenotypes, such as increased virus binding to α2–3 and α2–6 receptors (Appendix).
We conducted continuous phylogeographic analysis on this vulture-associated clade and revealed that nearly all (11 of 12) griffon vulture sequences clustered together in a distinct clade (Figure 2, panel A, node A). That clade is supported (posterior probability = 0.84) and contains vulture sequences from both Spain and France. Of note, the only other sequences in this clade originated from bearded vultures (Gypaetus barbatus) and a peregrine falcon (Falco peregrinus). This pattern suggests that the birds might have shared a common source of infection because of their scavenging feeding behaviors or that some birds could have been initially infected and subsequently transmitted the virus to others. The median time of the most recent common ancestor of the clade (Figure 3, panel A, node A) was estimated to be March 8, 2022 (95% highest posterior density [HPD] February 10, 2022–April 4, 2022). This clade was nested within a larger clade predominantly composed of wild bird sequences from Spain, including greylag geese (Anser anser), white storks (Ciconia ciconia), and grey herons (Ardea cinerea), but that also includes some poultry sequences, suggesting a likely wild bird origin with spillover events to both poultry and vultures (Figure 2, panel A).
Our continuous phylogeographic reconstruction indicated that the vulture viral lineages likely originated from central Spain (Figure 2, panel B) and substantial spatial dispersal was observed across 4 regions (Figure 2, panel C). That dispersal likely originated from wild birds in central Spain, as suggested by the predominance of wild bird sequences in the parent clade, with subsequent spillover events to both poultry and vultures (Figure 2, panel A) and >1 dispersal event toward Massif Central in early March (median date March 3, 2022; 95% HPD February 7, 2022–April 4, 2022; posterior probability = 0.92). Subsequently, lineages spread to the Pyrenees by the end of March (median date March 29, 2022 [95% HPD March 4, 2022–April 20, 2022]; posterior probability = 0.73) and to southern Spain by the end of April (median date April 21, 2022 [95% HPD March 28, 2022–May 4, 2022]; posterior probability = 0.44).
One griffon vulture sequence originating from a rehabilitation center in northern Spain (PP150341; site 6 in Figure 1) fell in a distinct position within the phylogenetic tree (Appendix Figure 2) within a clade of predominantly seabird sequences originating from France. This particular PP150341 sequence is positioned near the BB genotype (H5N1-A/Herring_gull/France/22P015977/2022-like), which has been rapidly expanding across Europe since 2022 (22).
Dispersal of HPAI by Griffon Vulture Movements
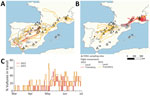
To examine changes in the movement patterns of griffon vultures during the HPAI outbreak, we focused on the 4-month period of March–June and compared the movements of flying birds during 2022 compared with 2023. From 114 vultures tagged before the start of the outbreak in Spain and France (94 adult and 20 immature birds, none of which were sampled for viral and serologic monitoring), the percentage of immature birds displaying long-range transit movements was significantly larger than the percentage of adult birds (88% immature vs. 24% adult when both years were pooled; χ2 = 37.01, degrees of freedom [d.f.] = 1; p<0.001). The proportion of long-range transit movements did not differ significantly between years (χ2 = 0.76, d.f. = 1, p = 0.268 for adults; χ2 = 0.09, p = 0.610 for immature birds).
We chose 16 vultures (4 immature and 4 adult birds in both 2022 and 2023) that best visually represent long-range transit movements to further investigate the spatial and temporal dimensions of those movements (Figure 3). The DDT were significantly longer for days of transit compared with days of local movements (181.8 + 77.4 km [max 438.2 km] for days of transit compared with 69.0 + 47.5 km [max 272.1 km] for days of local movement) (Appendix Table 8). Regardless of the type of movement, DDT increased significantly (p<0.001) and progressively over the course of the period (shortest in March and longest in June) (Figure 3, panel C). We observed no qualitative differences in transit movement patterns between 2022 and 2023. In transit movements, adults and immature birds traveled at similar speed, but in general adults took more direct trajectories (Figure 3, panel B). Overall, griffon vultures were able to travel between southern Spain and the Pyrenees or between the Pyrenees and the Alps in 1–2 days.
HPAI H5N1 infections were first detected in griffon vultures in southern Europe in spring 2022 in Spain and France. Infection led to central nervous clinical signs as well as reduced activity, immobility at the roost or nest, and death in some adults. Deaths were also recorded in nestlings, either from direct effects of infection or lack of parental care (19).
We investigated the origin of infection and assessed the nature and magnitude of viral spread through an integrated analysis of virologic, serologic, genomic, and ecologic data obtained from field sampling in France and Spain. In addition, we evaluated potential sources of exposure, studied population connectivity pathways, and investigated the potential of long-range contamination by movements of infected birds using long-term GPS tracking data.
Serologic results reflected the circulation of H5 HPAI among flying birds of all sampled colonies of griffon vultures. The high ELISA seroprevalence observed in flying birds seems to indicate widespread transmission but high survival within the meta-population, as opposed to the dramatic mortality rates observed in Sandwich terns, northern gannets, and bald eagles (20,23,24). In contrast, ELISA seroprevalence was considerably lower in nestlings; seropositivity was detected in just 1 colony (Appendix Table 1). That lower seroprevalence could be because of a reduced exposure of nestlings or the result of a high mortality rate. Those seropositive nestlings, detected at the same time as H5N1 AIV–mediated deaths of nestlings in neighboring nests, fledged successfully, demonstrating that infected nestlings, as well as infected adults, were able to survive the infection.
The spread of the virus through the griffon vulture populations in Spain and France was very fast; most H5 HPAI PCR–positive dead birds were collected during April–June 2022, and almost all live and dead birds sampled in both countries after that period tested negative for H5 HPAI virus by qRT-PCR. None of the antibody-positive vultures from our sample set tested positive by PCR, suggesting the absence of active HPAI H5N1 circulation after June 2022, which might relate to the clearance of infection by surviving vultures (25,26). Sandwich terns sampled in 2022 and 2023 showed a similar pattern; seropositivity was detected in adults in the absence of viral shedding (24). However, the lack of data regarding persistence of AIV immunity in griffon vultures, which is known to be highly dependent on the species and age of the host, as well as the subtype of virus, does not preclude circulation at a low prevalence or in the absence of clinical signs (27,28). Presence of H5-specific AIV antibodies as much as a year after the outbreak, albeit at low titers (Appendix Table 2), might be from antibody persistence or reexposure or from exposure to a different H5 AIV. In fact, AIV seropositivity of vultures in Spain to AIV other than H5 before 2022 provides circumstantial evidence that exposure of vultures to AIV could occur occasionally, which contrasts with the common assumption before the H5N1 outbreak that vultures were either not exposed or not susceptible to AIV infection.
Of note, HPAIV-related deaths of bearded vultures (G. barbatus) were reported concurrently to the griffon vulture outbreak (SM17). In contrast, no evidence of infection was found in 2 other cooccurring species of vultures, namely cinereous vultures (Aegypius monachus) and Egyptian vultures (Neophron percnopterus), which also regularly feed alongside griffon vultures (data not shown).
Phylogenetic and phylogeographic analyses of H5 genetic sequences obtained during the HPAI H5N1 outbreak offered key insights into the potential origins and transmission dynamics of viral lineages among griffon vultures. Of note, the inclusion of almost all griffon vultures within the same genetic cluster, despite having been sampled from geographically distant locations in Spain and France, suggests that they could have shared a common source of infection because of their scavenging feeding behavior, or that some could have been initially infected and subsequently transmitted the virus to others.
Those analyses enable us to draw hypotheses regarding the origin of the virus that infected vultures. This virus was genetically close to strains found in greylag geese and other wetland species, such as grey herons and white storks. Griffon vultures rarely forage in wetlands in France, but they do more often in southern Spain, where they can feed on livestock or wild mammal carcasses in marshes (i.e., potentially close to waterbirds) (Appendix Figure 5). Interactions with waterbirds might also have occurred at small waterbodies in southern Spain, where griffon vultures regularly bathe. Contacts could also have occurred in open urban landfills in northern Spain, where griffon vultures regularly feed alongside other wild bird species that are highly susceptible to the virus, such as gulls and white storks (29–32). Previous studies have shown potential transmission of low pathogenic avian influenza viruses between species frequenting the same landfills (33,34). Finally, contacts might involve opportunistic carnivorous mammals, such as red foxes (Vulpes vulpes), in which deaths from HPAI H5N1 virus have been reported throughout Europe (35). Uncertainties remain regarding the specific mechanisms of the griffon vultures’ contamination.
Our phylogenetic and phylogeographic analyses further suggest that the introduction of HPAI in griffon vultures from poultry farms seems unlikely. This conclusion is also supported by the behavior of griffon vultures, which do not typically visit poultry farm premises. Moreover, culled poultry from affected farms are discarded under strict biosecurity regulations, making the contact of griffon vultures with infected dead poultry unlikely. However, we cannot rule out that griffon vultures might have accessed inadequately discarded undiagnosed dead backyard poultry in some regions of Spain that could also have been consumed by gulls and storks, leading to the further spread and detection of the strain in both wild waterbirds and vultures (31,32).
The limited genetic diversity of the virus observed in griffon vulture populations, contrasting with the wide geographic distribution of infected birds, suggests that the virus spread in the southwestern Europe metapopulation through intraspecific contamination. The ecology of griffon vultures, especially their feeding behavior and their colonial nature, could explain this finding. Dense short-term aggregations during feeding on carcasses or at vulture feeding stations and dumpsites, where hundreds of individual birds congregate, make griffon vultures particularly vulnerable to airborne pathogen transmission (36,37). Subsequently, movements of infected birds over long distances could easily have contributed to virus dissemination to the whole population (38).
Phylogeographic reconstructions reveal a spatial dissemination pattern across 4 distinct regions, originating from central Spain, spreading to France in the Massif central and the Alps, and subsequently disseminating to the western part of the Pyrenees and southern Spain. This finding is coherent with the analysis of telemetric data, which show an overlap in the distributions of several GPS-tagged birds in Spain and France and long-range movements occurring between populations, particularly in spring, concurrent with the 2022 outbreak (Appendix). Such movements were also observed in other years (39), and it thus seems unlikely for them to have been triggered by the outbreak, as observed with northern gannets (23,40,41).
The nestling that was found seronegative and without clinical signs but tested positive for HPAI H5 in the vascular feather provides circumstantial evidence for shedding during the presymptomatic period, because shedding from feather follicles has recently been described as an efficient route of HPAI transmission (42). Experimental infection of red-legged partridges (Alectoris rufa) with an HPAI H7N1 virus evidenced an incubation period of 3 days with shedding from day 1 (43), whereas a similar approach evidenced a 5-day incubation period in falcons (Falco spp.) experimentally infected with HPAI H5N1 (44). Thus, under the hypothesis of viral incubation lasting >3 days, a griffon vulture infected in the Pyrenees would have enough time to reach southern Spain or the French Alps before showing clinical signs and reduced mobility (19). As an example, an immature vulture (Imm_FR_JOR) traveled from southern Portugal to the French Alps in 6 full days (Appendix).
The outbreak described in this study appeared to have had only a mild effect in terms of the conservation of griffon vultures. Compared with long-lived seabird populations in which a large proportion of adults died, mortality in griffon vultures mostly affected nestlings and only few adult birds; adult survival is the most sensitive demographic parameter in such a long-lived species (45). In addition, the outbreak struck the world’s largest population of griffon vultures, which could withstand such an ephemeral reduction in breeding success. However, even if griffon vulture populations seem able to overcome this HPAI outbreak, they face multiple threats on a global scale, particularly poisoning and persecution (46–48). The introduction and circulation of a new infectious pathogen could add additional pressure on the population. Unfortunately, the consequences of a population collapse of necrophagous birds could be catastrophic, especially from a sanitary point of view, in that longer persistence of the carcasses they eliminate would increase risk for pathogen persistence and spread in the environment (17,49).
Despite a likely limited epidemiologic role of griffon vultures in the circulation of HPAI in Spain and France, with very low permeability between griffon vulture populations and poultry farms, the infection of this new compartment raises multiple questions. In particular, this outbreak demonstrates the ability of this virus (and potentially other highly contagious pathogens) to spread rapidly through a population after a single introduction and shows that even a rare event has the potential for devastating effects.
In conclusion, the recent evolution of HPAI H5N1 has led to this pathogen being considered a severe concern for endangered bird species, especially those with colonial and scavenging behavior. Integrating the epidemiology of the virus with the ecology of the host species is key to a better understanding of outbreak dynamics and possible effects on wildlife conservation. More generally, implementing a multidisciplinary approach will be necessary to overcome these new challenges.
Dr. Hirschinger is a researcher at the National Veterinary School of Toulouse within the IHAP research unit. His work focuses on wildlife conservation, with a particular interest in the ecology and epidemiology of wildlife diseases and their effects on population dynamics and ecosystem health.
Acknowledgments
We thank the staff from LPO Grands Causses and LPO Occitanie for providing access to demographic data on griffon vulture populations in Grands Causses and helping in capturing vultures for tagging and collecting samples; particularly Lea Giraud, Thierry David, Robert Straughan, Matthieu Vaslin, Yves Roullaud, and Anna Terras. We thank the staff from Parc National des Cévennes et Parc National des Pyrénées, Parc Naturel Regional du Vercors, and the members of associations Vautours en Baronnies and Saiak for helping in capturing vultures for tagging and collecting samples, particularly Jocelyn Fonderflick, Jérôme Lafitte, Michel Clouet, Isabelle Rebours, Didier Peyrusqué, Nicolas Renous, Christian Tessier, Gael Foilleret, and Julien Traversier. We thank the staff from ENVT and CEFE for helping in collecting samples in the field and analyzing data, particularly Cécile Caubet, Laetitia Lebre, and Christophe de Franceschi. We acknowledge the Fundación para la Conservación del Quebrantahuesos FCQ, the regional government of Aragon and all organisms partaking in the LIFE PRO Quebrantahuesos for access to griffon vulture samples collected in 2022 and 2023. We thank all the stakeholders who made possible the epidemiological monitoring of these birds (including SAGIR network and OFB and “Diputaciones Forales” of the Spanish Basque Country). We thank the “Diputación Foral de Gipuzkoa” for the video of the vulture with symptoms. We also thank all submitting laboratories for sharing H5N1 genome sequences in the GISAID database (https://www.gisaid.org), and especially Azucena Sanchez from Spanish “Laboratorio Central de Veterinaria” for helping us with tracing the geographical origin of some sequences.
Telemetry study of Griffon vultures was authorized in the Programme Personnel 961, coordinated by O. Duriez, under the supervision of the French ringing centre, CRBPO, Paris. Viral and serologic samples collected benefitted from special permission 2022-s-11 signed by the Prefecture de Lozère and DREAL Occitanie on July 19, 2022. Tracking and monitoring of vultures in France is supported by OSU-OREME (SO Ecopop) and the long-term Studies in Ecology and Evolution (SEE-Life) program of the CNR, and with technical assistance of PLT staff from CEFE (Christophe de Franceschi). E.N.V.T. participated in the framework of the “Chaire de Biosécurité et Santé Aviaires,” funded by the Direction Générale de l’Alimentation, Ministère de l’Agriculture et de la Souveraineté Alimentaire, France. S.D. acknowledges support from the Fonds National de la Recherche Scientifique (F.R.S.-FNRS, Belgium; grant no. F.4515.22), from the Research Foundation–Flanders (Fonds voor Wetenschappelijk Onderzoek — Vlaanderen, FWO, Belgium; grant no. G098321N), and from the European Union Horizon 2020 projects MOOD (grant agreement no. 874850) and LEAPS (grant agreement no. 101094685). E.A. was supported by MCIN/AEI/10.13039/501100011033 grant nos. FJC2021-047885-I. The sampling and analyses carried out at NEIKER have been financed by MCIN/AEI/10.13039/501100011033 (grant no. PID2020-114060RR-C31) and by Basque Government (grant no. VIGIA-2200003). Analysis carried out at IREC was supported byMCIN/AEI/10.13039/501100011033 (grant no. PID2020-114060RR-C32). Monitoring of the GPS-tagged birds in Spain was funded by the Comunidad de Bardenas Reales de Navarra and the Projects RNM-1925 and P18-RT-1321 (Junta de Andalucía), and Ecotone Telemetry 2017-12-026.
U.H., J.L.G., O.D., and G.L.L. conceptualized the study. J.H., U.H., A.S., M.B., J.A.D., C.L.G.L., V.A., X.G., E.A., J.A.S.Z., A.C.A., S.M.M., J.T., S.P., P.O., A.V.D.W., and O.D. provided resources. J.H., U.H., A.S., C.G., G.C., M.B., J.A.D., C.L.G.L., M.W., V.A., X.G., L.D.P., S.D., S.M.M., and O.D. conducted the formal analysis. J.H., U.H., C.G., G.C., O.D., and G.L.L. wrote the original draft. J.H., U.H., C.G., M.B., J.A.D., L.D.P., S.D., E.A., J.A.S.Z., A.C.A., J.T., T.B., O.D., and G.L.L. reviewed and edited the manuscript. U.H., O.D., and G.L.L. supervised the research. U.H., M.B., J.A.D., J.A.S.Z., T.B., J.L.G., and O.D. acquired the funding.
References
- Graziosi G, Lupini C, Catelli E, Carnaccini S. Highly pathogenic avian influenza (HPAI) H5 clade 2.3.4.4b virus infection in birds and mammals. Animals (Basel). 2024;14:1372. DOIPubMedGoogle Scholar
- Ramey AM, Hill NJ, DeLiberto TJ, Gibbs SEJ, Camille Hopkins M, Lang AS, et al. Highly pathogenic avian influenza is an emerging disease threat to wild birds in North America. J Wildl Manage. 2022;86:
e22171 . DOIGoogle Scholar - Clough C. Underestimating wild bird deaths from avian influenza. Vet Rec. 2022;191:221–2. DOIPubMedGoogle Scholar
- Klaassen M, Wille M. The plight and role of wild birds in the current bird flu panzootic. Nat Ecol Evol. 2023;7:1541–2. DOIPubMedGoogle Scholar
- Plaza PI, Gamarra-Toledo V, Euguí JR, Lambertucci SA. Recent changes in patterns of mammal infection with highly pathogenic avian influenza A(H5N1) virus worldwide. Emerg Infect Dis. 2024;30:444–52. DOIPubMedGoogle Scholar
- Wille M, Klaassen M. No evidence for HPAI H5N1 2.3.4.4b incursion into Australia in 2022. Influenza Other Respir Viruses. 2023;17:
e13118 . DOIPubMedGoogle Scholar - Garg S, Reed C, Davis CT, Uyeki TM, Behravesh CB, Kniss K, et al. Outbreak of highly pathogenic avian influenza A(H5N1) viruses in U.S. dairy cattle and detection of two human cases—United States, 2024. MMWR Morb Mortal Wkly Rep. 2024;73:501–5. DOIPubMedGoogle Scholar
- Pohlmann A, King J, Fusaro A, Zecchin B, Banyard AC, Brown IH, et al. Has epizootic become enzootic? Evidence for a fundamental change in the infection dynamics of highly pathogenic avian influenza in Europe, 2021. mBio. 2022;13:
e0060922 .PubMedGoogle Scholar - Scoizec A, Niqueux E, Schmitz A, Grasland B, Palumbo L, Huneau-Salaün A, et al. New patterns for highly pathogenic avian influenza and adjustment of prevention, control and surveillance strategies: the example of France. Viruses. 2024;16:101. DOIPubMedGoogle Scholar
- World Health Organization. Joint FAO/WHO/WOAH preliminary assessment of recent influenza A(H5N1) viruses [cited 2025 Mar 12]. https://cdn.who.int/media/docs/default-source/global-influenza-programme/2024_04_23_fao-woah-who_h5n1_assessment.pdf
- Aznar I, Baldinelli F, Stoicescu A, Kohnle L; European Food Safety Authority (EFSA). Annual report on surveillance for avian influenza in poultry and wild birds in Member States of the European Union in 2021. EFSA J. 2022;20:
e07554 .PubMedGoogle Scholar - Molini U, Yabe J, Meki IK, Ouled Ahmed Ben Ali H, Settypalli TBK, Datta S, et al. Highly pathogenic avian influenza H5N1 virus outbreak among Cape cormorants (Phalacrocorax capensis) in Namibia, 2022. Emerg Microbes Infect. 2023;12:
2167610 . DOIPubMedGoogle Scholar - Alexandrou O, Malakou M, Catsadorakis G. The impact of avian influenza 2022 on Dalmatian pelicans was the worst ever wildlife disaster in Greece. Oryx. 2022;56:813–813. DOIGoogle Scholar
- Banyard AC, Lean FZX, Robinson C, Howie F, Tyler G, Nisbet C, et al. Detection of highly pathogenic avian influenza virus H5N1 clade 2.3.4.4b in great skuas: a species of conservation concern in Great Britain. Viruses. 2022;14:212. DOIPubMedGoogle Scholar
- Camphuysen K, Gear S. Great skuas and northern gannets on Foula, summer 2022—an unprecedented, H5N1 related massacre [cited 2023 Jan 27]. https://dataverse.nioz.nl/citation?persistentId=doi:10.25850/nioz/7b.b.gd
- Pohlmann A, Stejskal O, King J, Bouwhuis S, Packmor F, Ballstaedt E, et al. Mass mortality among colony-breeding seabirds in the German Wadden Sea in 2022 due to distinct genotypes of HPAIV H5N1 clade 2.3.4.4b. J Gen Virol. 2023;104. DOIPubMedGoogle Scholar
- Plaza PI, Blanco G, Lambertucci SA. Implications of bacterial, viral and mycotic microorganisms in vultures for wildlife conservation, ecosystem services and public health. Ibis. 2020;162:1109–24. DOIGoogle Scholar
- Ducatez MF, Tarnagda Z, Tahita MC, Sow A, de Landtsheer S, Londt BZ, et al. Genetic characterization of HPAI (H5N1) viruses from poultry and wild vultures, Burkina Faso. Emerg Infect Dis. 2007;13:611–3. DOIPubMedGoogle Scholar
- Duriez O, Sassi Y, Le Gall-Ladevèze C, Giraud L, Straughan R, Dauverné L, et al. Highly pathogenic avian influenza affects vultures’ movements and breeding output. Curr Biol. 2023;33:3766–3774.e3. DOIPubMedGoogle Scholar
- Nemeth NM, Ruder MG, Poulson RL, Sargent R, Breeding S, Evans MN, et al. Bald eagle mortality and nest failure due to clade 2.3.4.4 highly pathogenic H5N1 influenza A virus. Sci Rep. 2023;13:191. DOIPubMedGoogle Scholar
- Fusaro A, Zecchin B, Giussani E, Palumbo E, Agüero-García M, Bachofen C, et al. High pathogenic avian influenza A(H5) viruses of clade 2.3.4.4b in Europe—why trends of virus evolution are more difficult to predict. Virus Evol. 2024;10:veae027. DOIPubMedGoogle Scholar
- Adlhoch C, Fusaro A, Gonzales JL, Kuiken T, Mirinaviciute G, Niqueux É, et al.; European Food Safety Authority, European Centre for Disease Prevention and Control, European Union Reference Laboratory for Avian Influenza. Avian influenza overview March - April 2023. EFSA J. 2023;21:
e08039 .PubMedGoogle Scholar - Jeglinski JWE, Lane JV, Votier SC, Furness RW, Hamer KC, McCafferty DJ, et al. HPAIV outbreak triggers short-term colony connectivity in a seabird metapopulation. Sci Rep. 2024;14:3126. DOIPubMedGoogle Scholar
- Knief U, Bregnballe T, Alfarwi I, Ballmann MZ, Brenninkmeijer A, Bzoma S, et al. Highly pathogenic avian influenza causes mass mortality in Sandwich Tern Thalasseus sandvicensis breeding colonies across north-western Europe. Bird Conserv Int. 2024;34:
e6 . DOIGoogle Scholar - Root JJ, Bosco-Lauth AM, Marlenee NL, Bowen RA. Viral shedding of clade 2.3.4.4 H5 highly pathogenic avian influenza A viruses by American robins. Transbound Emerg Dis. 2018;65:1823–7. DOIPubMedGoogle Scholar
- Bosco-Lauth AM, Marlenee NL, Hartwig AE, Bowen RA, Root JJ. Shedding of clade 2.3.4.4 H5N8 and H5N2 highly pathogenic avian influenza viruses in peridomestic wild birds in the U.S. Transbound Emerg Dis. 2019;66:1301–5. DOIPubMedGoogle Scholar
- Ellis JW, Root JJ, McCurdy LM, Bentler KT, Barrett NL, VanDalen KK, et al. Avian influenza A virus susceptibility, infection, transmission, and antibody kinetics in European starlings. PLoS Pathog. 2021;17:
e1009879 . DOIPubMedGoogle Scholar - Tolf C, Latorre-Margalef N, Wille M, Bengtsson D, Gunnarsson G, Grosbois V, et al. Individual variation in influenza A virus infection histories and long-term immune responses in mallards. PLoS One. 2013;8:
e61201 . DOIPubMedGoogle Scholar - Arévalo-Ayala DJ, Real J, Durà C, Aymerich J, Hernández-Matías A. Reduction of organic waste in a landfill lowers the visitation probability but not the local abundance of a long-lived scavenger species. Bird Conserv Int. 2023;33:
e15 . DOIGoogle Scholar - Rasmussen EA, Czaja A, Cuthbert FJ, Tan GS, Lemey P, Nelson MI, et al. Influenza A viruses in gulls in landfills and freshwater habitats in Minnesota, United States. Front Genet. 2023;14:
1172048 . DOIPubMedGoogle Scholar - Arrondo E, Sebastián-González E, Moleón M, Morales-Reyes Z, María Gil-Sánchez J, Cortés-Avizanda A, et al. Vulture culture: dietary specialization of an obligate scavenger. Proc Biol Sci. 2023;290:
20221951 .PubMedGoogle Scholar - Fernández-Gómez L, Cortés-Avizanda A, Arrondo E, García-Alfonso M, Ceballos O, Montelío E, et al. Vultures feeding on the dark side: current sanitary regulations may not be enough. Bird Conserv Int. 2022;32:590–608. DOIGoogle Scholar
- Bárbara A, Torrontegi O, Camacho MC, Barral M, Hernández JM, Höfle U. Avian influenza virus surveillance in south-central Spain using fecal samples of aquatic birds foraging at landfills. Front Vet Sci. 2017;4:178. DOIPubMedGoogle Scholar
- López-Calderón C, Martín-Vélez V, Blas J, Höfle U, Sánchez MI, Flack A, et al. White stork movements reveal the ecological connectivity between landfills and different habitats. Mov Ecol. 2023;11:18. DOIPubMedGoogle Scholar
- Lagan P, McKenna R, Baleed S, Hanna B, Barley J, McConnell S, et al. Highly pathogenic avian influenza A(H5N1) virus infection in foxes with PB2-M535I identified as a novel mammalian adaptation, Northern Ireland, July 2023. Euro Surveill. 2023;28:
2300526 . DOIPubMedGoogle Scholar - Ellis J, Shriner S, McLean H, Petersen L, Root JJ. Inventory of wildlife use of mortality pits as feeding sites: implications of pathogen exposure [cited 2023 Jan 31]. https://digitalcommons.usu.edu/hwi/vol11/iss1/4/
- Bosè M, Duriez O, Sarrazin F. Intra-specific competition in foraging Griffon Vultures Gyps fulvus: 1. Dynamics of group feeding. Bird Study. 2012;59:182–92. DOIGoogle Scholar
- Fluhr J, Benhamou S, Peyrusque D, Duriez O. Space use and time budget in two populations of griffon vultures in contrasting landscapes. J Raptor Res. 2021;55:425–37. DOIGoogle Scholar
- Delgado-González A, Cortés-Avizanda A, Serrano D, Arrondo E, Duriez O, Margalida A, et al. Apex scavengers from different European populations converge at threatened savannah landscapes. Sci Rep. 2022;12:2500. DOIPubMedGoogle Scholar
- Boulinier T. Avian influenza spread and seabird movements between colonies. Trends Ecol Evol. 2023;38:391–5. DOIPubMedGoogle Scholar
- Careen NG, Collins SM, D’Entremont KJN, Wight J, Rahman I, Hargan KE, et al. Highly pathogenic avian influenza virus resulted in unprecedented reproductive failure and movement behaviour by northern gannets. Mar Ornithol. 2024;52:121–8.
- Gaide N, Filaire F, Bertran K, Crispo M, Dirat M, Secula A, et al. The feather epithelium contributes to the dissemination and ecology of clade 2.3.4.4b H5 high pathogenicity avian influenza viruses in ducks. Emerg Microbes Infect. 2023;12:
2272644 . DOIPubMedGoogle Scholar - Bertran K, Pérez-Ramírez E, Busquets N, Dolz R, Ramis A, Darji A, et al. Pathogenesis and transmissibility of highly (H7N1) and low (H7N9) pathogenic avian influenza virus infection in red-legged partridge (Alectoris rufa). Vet Res (Faisalabad). 2011;42:24. DOIPubMedGoogle Scholar
- Bertran K, Busquets N, Abad FX, García de la Fuente J, Solanes D, Cordón I, et al. Highly (H5N1) and low (H7N2) pathogenic avian influenza virus infection in falcons via nasochoanal route and ingestion of experimentally infected prey. PLoS One. 2012;7:
e32107 . DOIPubMedGoogle Scholar - Robert A, Sarrazin F, Couvet D, Legendre S. Releasing adults versus young in reintroductions: interactions between demography and genetics. Conserv Biol. 2004;18:1078–87. DOIGoogle Scholar
- Ogada DL, Keesing F, Virani MZ. Dropping dead: causes and consequences of vulture population declines worldwide. Ann N Y Acad Sci. 2012;1249:57–71. DOIPubMedGoogle Scholar
- Buechley ER, Şekercioğlu ÇH. The avian scavenger crisis: looming extinctions, trophic cascades, and loss of critical ecosystem functions. Biol Conserv. 2016;198:220–8. DOIGoogle Scholar
- Posillico M, Costanzo A, Bottoni S, Altea T, Opramolla G, Pascazi A, et al. Reported mortality of griffon vulture Gyps fulvus in central Italy and indications for conservation and management. Bird Conserv Int. 2023;33:
e68 . DOIGoogle Scholar - Ogada DL, Torchin ME, Kinnaird MF, Ezenwa VO. Effects of vulture declines on facultative scavengers and potential implications for mammalian disease transmission. Conserv Biol. 2012;26:453–60. DOIPubMedGoogle Scholar
Figures
Table
Cite This ArticleOriginal Publication Date: July 16, 2025
Table of Contents – Volume 31, Number 8—August 2025
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Guillaume Le Loc’h, UMR 1225 IHAP, École Nationale Vétérinaire de Toulouse, 23 Chemin des Capelles, 31300 Toulouse, France
Top