Recombinant Myxoma Virus in European Brown Hares, 2023–2024
Luisa Fischer

, Erwin de Bruin, Evelien Jongepier, Erik Koffeman, Patricia König, Florian Pfaff, Martin Peters, Judith M.A. van den Brand, Marin Bussi, Dominik Fischer, Valentina Caliendo, Erik Weerts, Jooske IJzer, Janina Müller, Ann-Katrin Kühling, Maren Kummerfeld, Jana Müller, Henning Petersen, Sabine Merbach, Martin Beer, Mathilde Uiterwijk, Karst de Boer, and Jolianne M. Rijks
Author affiliation: Research Center for Hunting Science and Wildlife Management, State Agency for Consumer Protection and Nutrition North Rhine-Westphalia, Bonn, Germany (L. Fischer); Dutch Wildlife Health Centre, Utrecht University, Utrecht, the Netherlands (E. de Bruin, J.M.A. van den Brand, M. Bussi, V. Caliendo, E. Weerts, J. IJzer, J.M. Rijks); Royal Dutch Hunters’ Association, Amersfoort, the Netherlands (E. Jongepier); Fauna Management Unit Gelderland, Arnhem, the Netherlands (E. Koffeman); Friedrich-Loeffler-Institut, Greifswald-Insel Riems, Germany (P. König, F. Pfaff, M. Beer); Chemical and Veterinary Investigation Office Westphalia, Arnsberg, Germany (M. Peters, S. Merbach); Der Grüne Zoo Wuppertal, Wuppertal, Germany (D. Fischer); Chemical and Veterinary Investigation Office Rhein-Ruhr-Wupper, Krefeld, Germany (Janina Müller, A.-K. Kühling); Chemical and Veterinary Investigation Office Münsterland-Emscher-Lippe, Münster, Germany (M. Kummerfeld); Chemical and Veterinary Investigation Office Ostwestfalen-Lippe, Detmold, Germany (Jana Müller, H. Petersen); Centre for Monitoring of Vectors, Netherlands Food and Consumer Product Safety Authority, Wageningen, the Netherlands (M. Uiterwijk, K. de Boer)
Main Article
Figure 3
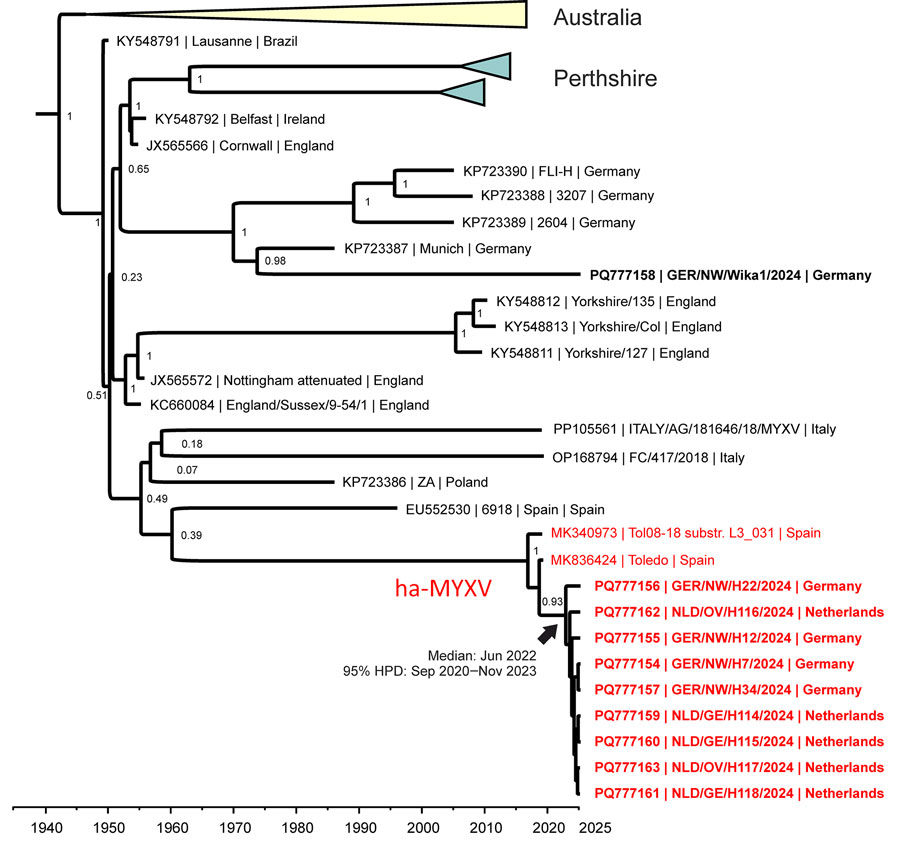
Figure 3. Time-based phylogeny of MYXV from a 2024 outbreak in European brown hares (Lepus europaeus) in the border area of the Netherlands and Germany and reference sequences. Ten full-length MYXV sequences from the outbreak were aligned to 114 available full-length MYXV reference genome sequences from GenBank and used for time-based phylogenetic analyses with BEAST version 1.10.4 (https://github.com/beast-dev/beast-mcmc/releases/tag/v1.10.4). Red indicates isolates belonging to ha-MYXV; bold text indicates sequences from this study. Branch labels represent statistical support values; values closer to 1 indicate stronger support. GenBank accession numbers are shown. ha-MYXV, hare-adapted natural recombinant MYXV; HPD, highest posterior density; MYXV, myxoma virus.
Main Article
Page created: July 02, 2025
Page updated: July 22, 2025
Page reviewed: July 22, 2025
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
