Volume 31, Number 8—August 2025
Dispatch
Identification of Co-Circulating Dengue and South America–Origin Zika Viruses, Pakistan, 2021–2022
Figure
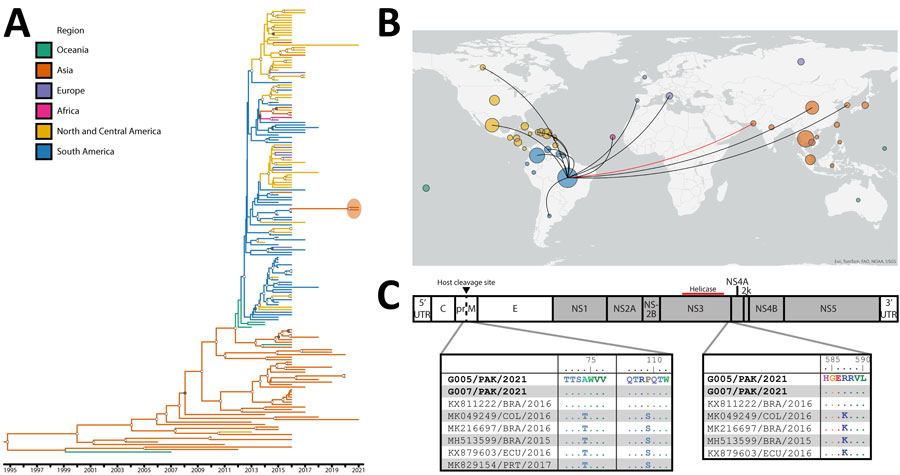
Figure. Phylogenetic analysis of Zika viruses (ZIKV) identified in a study of co-circulation of dengue and South American–origin Zika viruses, Pakistan, 2021–2022. A) BEAST (https://beast.community/index.html) time-aware maximum clade credibility tree describing inferred genetic lineage of global Asian-lineage ZIKV, colored by observed and estimated geographic origin. Branch backbones are colored when called with >80% confidence by Augur. Two newly described viruses from Pakistan are circled in orange, with the closest observed ancestors derived from circulating South American ZIKV. Open circles indicate posterior probability >0.9, solid dots 0.8–0.9. B) Phylogeographic map illustrating inferred international ZIKV transmission events originating in Brazil from viruses included in panel A. Map was visualized by inferred origin and transmission using Auspice (https://docs.nextstrain.org/projects/auspice/en/stable/index.html). Circle size is relative to the number of included viruses from the country and colored by continent. Red line highlights the inferred Brazil to Pakistan incursion. C) ZIKV sequences selected from the same clade and subclade as Pakistan-origin viruses aligned to G005/PAK/2021. Identical amino acid residues are shown as dots. We identified 2 changes in prM, T74A and S109P (left), and 1 in NS3, K587R (right). G005/PAK/2021 corresponds to patient E; G007/PAK/2021 corresponds to patient F. NS, nonstructural; UTR, untranslated region.
1These first authors contributed equally to this article.