Volume 31, Number 8—August 2025
Perspective
A Roadmap of Primary Pandemic Prevention Through Spillover Investigation
Cite This Article
Citation for Media
Abstract
Since the COVID-19 pandemic, attention and investment in pandemic preparedness have increased. Although there are many valiant plans around pandemic preparedness, they typically involve slowing the spread or mitigating the effects of a pathogen after it has already entered the human population. The task of stopping the pathogen from entering the human population in the first place, spillover prevention, remains a neglected area in discussions and planning for pandemic risk mitigation. Every spillover offers an opportunity to learn about an emerging public health threat and the conditions that aligned to enable spillover occurrence. In this article, we outline One Health approaches for use in spillover investigations, drawing from our experience investigating Hendra and Nipah virus spillovers. We present a roadmap for how findings from those investigations can lead to the development of interventions for spillover and ultimately pandemic prevention.
Pandemics occur when a pathogen is transmitted across continents through human populations that lack prior immunity (1). Most pathogens that start pandemics are zoonotic, originating in wildlife or other animals (1). Typically, those animal pathogens are novel to humans, so most humans are susceptible, and if those pathogens have or gain the ability to transmit between humans, they pose a pandemic risk. In the wake of the COVID-19 pandemic, pandemic preparedness has been a focus of global engagement. Although such efforts include valiant plans, they largely focus on slowing the spread or mitigating the effects of a pathogen after it has already entered the human population. Initiatives of note include the Coalition for Epidemic Preparedness Innovations plans to deliver vaccines within 100 days of an emerging threat, the World Bank’s investment in surveillance in low- and middle-income countries, and the World Health Organization’s efforts to develop more rigorous global agreements on investigation and collective action. Although those strategies enhance our responses to emerging infection outbreaks, they primarily address scenarios after a pathogen has established transmission between humans. However, the task of stopping the pathogen from entering the human population in the first place, spillover prevention, remains a neglected area in discussions and plans for pandemic risk mitigation.
A spillover occurs when a pathogen infects a new host species (2,3). The vast majority of spillovers will not lead to an outbreak or pandemic. However, for pathogens with pandemic potential, each spillover into a human is an opportunity to launch a pandemic. Most pandemic prevention plans focus on finding outbreak events earlier, notifying neighboring countries, assembling effective outbreak response teams, and enhancing global surveillance for spillover and outbreak events. Those measures are all crucial. However, preventing the spillover in the first place should be a fundamental component of our global strategy for preventing pandemics.
Numerous initiatives have attempted to identify potential pandemic causing pathogens before they cause outbreaks. One approach is to model geographic areas at high risk for spillovers, correlating putative drivers with locations of past spillovers and overlap of humans and reservoir species (4–6). Those efforts aim to focus surveillance and resources on areas or species of high risk. Substantial investments have led to the discovery of new viruses infecting rodents, bats, and primates, including viruses that were phylogenetically related to outbreak causing pathogens, suggesting a potential risk for spillover (7–19). Although such efforts have produced findings of interest, they have not produced actionable public health data. Those approaches do not inform which pathogens are spilling over and the mechanisms driving these events.
Spillovers do provide actionable data. Once an emerging pathogen infects a human, a public health threat is actualized. Those events garner our attention and concern much more than hypothetical risk warnings. Particularly alarming is evidence of transmission of the pathogen from human to human, because this capability is necessary to cause a pandemic. For example, if there was evidence that persons infected with bovine strains of avian influenza H5 across the United States (20) were able to infect others, the risk of a pandemic from this virus would increase drastically.
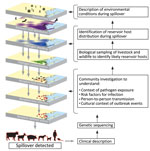
Every spillover offers a critical opportunity to learn about an emerging public health threat and the conditions that aligned to enable the spillover occurrence. Investigating those events requires a transdisciplinary approach, often best conceptualized as a One Health investigation that integrates multiple fields of expertise (Figure 1). The investigation typically begins with medical experts who understand the clinical manifestations of the disease and natural history of infection because the spillover is detected when a sick person seeks care. Spillovers sometimes also occur first in other species, which become bridging hosts to humans. Laboratory analysis of the genetic sequence of the pathogen can provide more information about its origins and potential reservoir hosts. Concurrently, epidemiologic investigations can determine the exposures that led to infection and assess if transmission is ongoing through extensive contact tracing efforts. Next, veterinary and ecologic investigations of animals in the affected communities can identify potential reservoir species and bridging hosts. Social scientists contribute in-depth understanding of how local practices might have enabled exposure and transmission, including human–animal interactions and their drivers. Finally, environmental and ecologic investigations elucidate how changes in the reservoir host condition or distribution might have enabled spillover. The timing of those investigations is critical because the conditions for spillover can be fleeting, so rapid identification and investigation of spillovers is vital.
One Health spillover investigations represent a crucial step in a broader continuum of actions designed to move from identifying mechanistic, proximal causes of spillover to designing and testing interventions to prevent them. This continuum from discovery to spillover prevention (Figure 2) encompasses multiple interconnected steps: discovery of the zoonotic pathogen in reservoir hosts, detection of spillover events, carrying out One Health spillover investigations, and identifying the transmission pathways and conditions that enabled spillover. The subsequent steps involve iterative research to develop, test, and deploy interventions to prevent spillovers by targeting both proximal and upstream causes. Each step informs the others, creating ongoing feedback essential for pandemic prevention.
Spillover investigations are crucial for pandemic prevention, and more effort is needed to identify and study spillovers. There are multiple barriers to identifying spillovers that span global, national, and local levels. Because of those barriers, many spillovers remain undetected or unreported. At the local level, there might be insufficient resources to diagnose common causes of disease, much less rare and emerging pathogens. Even if that barrier is overcome, communities might be apprehensive about uncovering emerging pathogens because that process can lead to blame, stigmatization, and negative economic impacts. At the national level, there are political, financial, and economic threats to navigate. The reality is that spillovers are almost always negative events for governments. Spillovers are politically sensitive and sometimes not reported out of fear. Reporting of emerging pathogen outbreaks has led to severe economic outcomes for reporting countries, including travel bans or trade embargoes (21–23). Once a spillover is identified, governments might be expected to expend considerable resources for investigation and response to reduce the global pandemic risk. For governments that have threadbare budgets for combating endemic public health problems, there might be little desire to take on those additional actions. Although the numerous disincentives to spillover detection are formidable, we have much to gain by overcoming them.
When investigations of spillovers, particularly those conducted through a One Health approach, have taken place, they have yielded critical insights and even solutions to prevent future spillovers (Figure 2). For example, Hendra virus is an often-fatal virus transmitted from bats to horses and subsequently to humans in Australia. Ecologists involved in the investigations of Hendra virus spillovers noted unusual bat activity in the paddocks of affected horses. Bats were feeding on unripe figs and other foods associated with starvation avoidance. This observation prompted the researchers to hypothesize that food shortages for bats were somehow associated with spillovers. Subsequent long-term studies revealed that climate fluctuations, interacting with habitat loss, led to acute food shortages that drove bats into agricultural areas and caused them to shed Hendra virus in proximity to horses (24). During those investigations, researchers noted that spillovers did not occur when remnant patches of critical habitat flowered, providing food for bats. This finding suggested a potential solution: restoring critical habitats to mitigate spillovers (24). This example illustrates the critical role of spillover investigation and subsequent studies to understand the mechanisms underlying spillovers. When mechanisms are understood, interventions to prevent future occurrences become apparent. Restoration of critical habitat has begun, but it will take more than a decade to determine if that intervention decreases the risk for Hendra virus spillovers.
Nipah virus transmission in Bangladesh provides another excellent case study about how looking for spillovers and then conducting One Health investigations have led to major insights into proximal causes of spillover and possible targets for spillover prevention (Figure 2). The first outbreaks of Nipah virus were discovered in Bangladesh in 2001 (25), and after years of One Health investigations of spillovers, an understanding of the source began to form in 2005 (26). Epidemiologic studies identified date palm sap consumption as a key risk factor for Nipah virus infection, and social scientists studied how the sap was harvested and sold (26–29). Date palm sap is collected from trees and drunk fresh during the cool, winter months; it is a cultural delicacy (29). Wildlife investigations identified that bats shed virus in their urine and saliva (30), ecologic investigations revealed that bats drink and contaminate date palm sap as it drips into the pots (31), and virologic studies showed that Nipah virus is stable in date palm sap (32). Further studies then demonstrated that simple covers of the pots and sap stream on the tree, which were already being used by some sap collectors, would protect the sap from contact with bats (33,34).
Spillover dynamics are driven by the interaction of multiple complex systems, including infection dynamics in the reservoir hosts, their shifting population distributions, and emergent human behaviors and practices (Figure 1). Drivers span from local alterations in land use change to global climate. Investigating the underlying drivers of spillovers often requires sustained effort over years or decades (Figure 1), extending beyond the duration of individual grants, or any single person’s tenure in a particular job. However, the example of Hendra virus spillover investigations in Australia exemplifies how a strong curiosity and a commitment to understanding the mechanisms underlying spillovers can lead to the potential for ecological solutions to prevent pandemics (24).
In summary, we have presented evidence about how a One Health approach to spillover investigation can lead to spillover prevention by using Hendra and Nipah virus as case studies. However, those approaches are applicable to any spillover pathogen, not just viruses, and any reservoir host, not just bats. Opportunities to learn more about and prevent spillovers are frequent but often missed. We know very little about the specific spillovers that led to most of the large outbreaks or pandemics in the past 100 years, mostly because by the time investigations began, the trail was cold. For example, the origin of the 2013–2016 Ebola epidemic in West Africa was not investigated until months after its onset, leaving the initial spillover that led to that outbreak uncertain, similar to most other Ebola outbreaks (35,36). The origins of several recent spillovers remain unresolved, including how Nipah virus first spilled over to humans in Kerala, India, in 2018, 2019, and 2023 (37,38), and how H5N1 spilled over into dairy cattle in the United States (20). The origins of the COVID-19 pandemic are likely to remain unsolved indefinitely, because of delays in investigations. Until we dedicate ourselves to the search for and One Health investigation of spillovers, we remain vulnerable to their devastating consequences.
Dr. Gurley is an epidemiologist at the Johns Hopkins Bloomberg School of Public Health. Her research interests include the mechanisms of zoonotic spillover and prevention strategies for Nipah virus and other emerging infections in Bangladesh.
Dr. Plowright is a professor at Cornell University and a Cornell Atkinson Scholar at the Cornell Atkinson Center for Sustainability. Her research interests include transdisciplinary approaches to identify the mechanisms driving zoonotic spillover and informing strategies to prevent the emergence of novel pathogens and future pandemics.
Acknowledgment
This study was funded in part by the US National Science Foundation (grant no. EF-2133763/EF-2231624) and the Defense Advanced Research Projects Agency PREempt Emerging Pathogenic Threats program (cooperative agreement no. D18AC00031). The content of the information does not necessarily reflect the position or the policy of the US government, and no official endorsement should be inferred.
References
- Dias RA. Towards a comprehensive definition of pandemics and strategies for prevention: a historical review and future perspectives. Microorganisms. 2024;12:1802. DOIPubMedGoogle Scholar
- Plowright RK, Parrish CR, McCallum H, Hudson PJ, Ko AI, Graham AL, et al. Pathways to zoonotic spillover. Nat Rev Microbiol. 2017;15:502–10. DOIPubMedGoogle Scholar
- Lloyd-Smith JO, George D, Pepin KM, Pitzer VE, Pulliam JR, Dobson AP, et al. Epidemic dynamics at the human-animal interface. Science. 2009;326:1362–7. DOIPubMedGoogle Scholar
- Pigott DM, Golding N, Mylne A, Huang Z, Henry AJ, Weiss DJ, et al. Mapping the zoonotic niche of Ebola virus disease in Africa. Elife. 2014;3:
e04395 . DOIPubMedGoogle Scholar - Muylaert RL, Wilkinson DA, Kingston T, D’Odorico P, Rulli MC, Galli N, et al. Using drivers and transmission pathways to identify SARS-like coronavirus spillover risk hotspots. Nat Commun. 2023;14:6854. DOIPubMedGoogle Scholar
- Han BA, Schmidt JP, Bowden SE, Drake JM. Rodent reservoirs of future zoonotic diseases. Proc Natl Acad Sci U S A. 2015;112:7039–44. DOIPubMedGoogle Scholar
- Goldstein T, Anthony SJ, Gbakima A, Bird B, Bangura J, Tremeau-Bravard A, et al. The discovery of a new Ebolavirus, Bombali virus, adds further support for bats as hosts of Ebolaviruses. Int J Infect Dis. 2019;79:4–5. DOIGoogle Scholar
- Valitutto MT, Aung O, Tun KYN, Vodzak ME, Zimmerman D, Yu JH, et al. Detection of novel coronaviruses in bats in Myanmar. PLoS One. 2020;15:
e0230802 . DOIPubMedGoogle Scholar - Cameron K, Hayes B, Olson SH, Smith BR, Pante J, Laudisoit A, et al. Detection of first gammaherpesvirus sequences in Central African bats. New Microbes New Infect. 2020;36:
100705 . DOIPubMedGoogle Scholar - Amman BR, Bird BH, Bakarr IA, Bangura J, Schuh AJ, Johnny J, et al. Isolation of Angola-like Marburg virus from Egyptian rousette bats from West Africa. Nat Commun. 2020;11:510. DOIPubMedGoogle Scholar
- Nziza J, Goldstein T, Cranfield M, Webala P, Nsengimana O, Nyatanyi T, et al. Coronaviruses detected in bats in close contact with humans in Rwanda. Ecohealth. 2020;17:152–9. DOIPubMedGoogle Scholar
- Kumakamba C, N’Kawa F, Kingebeni PM, Losoma JA, Lukusa IN, Muyembe F, et al. Analysis of adenovirus DNA detected in rodent species from the Democratic Republic of the Congo indicates potentially novel adenovirus types. New Microbes New Infect. 2019;34:
100640 . DOIPubMedGoogle Scholar - Lange CE, Niama FR, Cameron K, Olson SH, Aime Nina R, Ondzie A, et al. First evidence of a new simian adenovirus clustering with Human mastadenovirus F viruses. Virol J. 2019;16:147. DOIPubMedGoogle Scholar
- Diffo J, Ndze VN, Ntumvi NF, Takuo JM, Mouiche MMM, Tamoufe U, et al. DNA of diverse adenoviruses detected in Cameroonian rodent and shrew species. Arch Virol. 2019;164:2359–66. DOIPubMedGoogle Scholar
- Islam A, Islam S, Ferdous J, Rahman MK, Uddin MH, Akter S, et al. Diversity and prevalence of parasitic infestation with zoonotic potential in dromedary camel ( Camelus dromedarius ) and fat-tailed sheep (dhumba) in Bangladesh. J Adv Vet Anim Res. 2019;6:142–7. DOIPubMedGoogle Scholar
- Lacroix A, Duong V, Hul V, San S, Davun H, Omaliss K, et al. Genetic diversity of coronaviruses in bats in Lao PDR and Cambodia. Infect Genet Evol. 2017;48:10–8. DOIPubMedGoogle Scholar
- Epstein JH, Anthony SJ. Viral discovery as a tool for pandemic preparedness. Rev Sci Tech. 2017;36:499–512. DOIPubMedGoogle Scholar
- Yang X-L, Zhang Y-Z, Jiang R-D, Guo H, Zhang W, Li B, et al. Genetically diverse filoviruses in Rousettus and Eonycteris spp. bats, China, 2009 and 2015. Emerg Infect Dis. 2017;23:482–6. DOIPubMedGoogle Scholar
- Ge X-Y, Yang W-H, Pan H, Zhou JH, Han X, Zhu GJ, et al. Fugong virus, a novel hantavirus harbored by the small oriental vole (Eothenomys eleusis) in China. Virol J. 2016;13:27. DOIPubMedGoogle Scholar
- Centers for Disease Control and Prevention. H5 bird flu: current situation. Avian influenza (bird flu). 2025 [cited 2025 Feb 17]. https://www.cdc.gov/bird-flu/situation-summary/index.html
- Pattani R. Unsanctioned travel restrictions related to Ebola unravel the global social contract. CMAJ. 2015;187:166–7. DOIPubMedGoogle Scholar
- Meier BM, Bueno de Mesquita J, Burci GL, Chirwa D, Dagron S, Eccleston-Turner M, et al. Travel restrictions and variants of concern: global health laws need to reflect evidence. Bull World Health Organ. 2022;100:178–178A. DOIPubMedGoogle Scholar
- Worsnop CZ. Domestic politics and the WHO’s International Health Regulations: Explaining the use of trade and travel barriers during disease outbreaks. Rev Int Organ. 2017;12:365–95. DOIPubMedGoogle Scholar
- Eby P, Peel AJ, Hoegh A, Madden W, Giles JR, Hudson PJ, et al. Pathogen spillover driven by rapid changes in bat ecology. Nature. 2023;613:340–4. DOIPubMedGoogle Scholar
- Hsu VP, Hossain MJ, Parashar UD, Ali MM, Ksiazek TG, Kuzmin I, et al. Nipah virus encephalitis reemergence, Bangladesh. Emerg Infect Dis. 2004;10:2082–7. DOIPubMedGoogle Scholar
- Luby SP, Rahman M, Hossain MJ, Blum LS, Husain MM, Gurley E, et al. Foodborne transmission of Nipah virus, Bangladesh. Emerg Infect Dis. 2006;12:1888–94. DOIPubMedGoogle Scholar
- Hegde ST, Sazzad HMS, Hossain MJ, Alam MU, Kenah E, Daszak P, et al. Investigating rare risk factors for Nipah virus in Bangladesh: 2001–2012. Ecohealth. 2016;13:720–8. DOIPubMedGoogle Scholar
- Chakraborty A, Sazzad HMS, Hossain MJ, Islam MS, Parveen S, Husain M, et al. Evolving epidemiology of Nipah virus infection in Bangladesh: evidence from outbreaks during 2010-2011. Epidemiol Infect. 2016;144:371–80. DOIPubMedGoogle Scholar
- Nahar N, Sultana R, Gurley ES, Hossain MJ, Luby SP. Date palm sap collection: exploring opportunities to prevent Nipah transmission. Ecohealth. 2010;7:196–203. DOIPubMedGoogle Scholar
- Anderson DE, Islam A, Crameri G, Todd S, Islam A, Khan SU, et al. Isolation and full-genome characterization of Nipah viruses from bats, Bangladesh. Emerg Infect Dis. 2019;25:166–70. DOIPubMedGoogle Scholar
- Khan MSU, Hossain J, Gurley ES, Nahar N, Sultana R. Use of infrared camera to understand bats’ access to date palm sap: implications for preventing Nipah virus transmission. Ecohealth 2010. https://link.springer.com/article/10.1007/s10393-010-0366-2.
- Fogarty R, Halpin K, Hyatt AD, Daszak P, Mungall BA. Henipavirus susceptibility to environmental variables. Virus Res. 2008;132:140–4. DOIPubMedGoogle Scholar
- Nahar N, Mondal UK, Sultana R, Hossain MJ, Khan MS, Gurley ES, et al. Piloting the use of indigenous methods to prevent Nipah virus infection by interrupting bats’ access to date palm sap in Bangladesh. Health Promot Int. 2013;28:378–86. DOIPubMedGoogle Scholar
- Khan SU, Gurley ES, Hossain MJ, Nahar N, Sharker MAY, Luby SP. A randomized controlled trial of interventions to impede date palm sap contamination by bats to prevent nipah virus transmission in Bangladesh. PLoS One. 2012;7:
e42689 . DOIPubMedGoogle Scholar - Marí Saéz A, Weiss S, Nowak K, Lapeyre V, Zimmermann F, Düx A, et al. Investigating the zoonotic origin of the West African Ebola epidemic. EMBO Mol Med. 2015;7:17–23. DOIPubMedGoogle Scholar
- Plowright RK, Hudson PJ. From protein to pandemic: the transdisciplinary approach needed to prevent spillover and the next pandemic. Viruses. 2021;13:1298. DOIPubMedGoogle Scholar
- Arunkumar G, Chandni R, Mourya DT, Singh SK, Sadanandan R, Sudan P, et al.; Nipah Investigators People and Health Study Group. Outbreak investigation of Nipah virus disease in Kerala, India, 2018. J Infect Dis. 2019;219:1867–78. DOIPubMedGoogle Scholar
- Rahim AA, Chandran P, Bindu V, Radhakrishnan C, Moorkoth AP, Ramakrishnan LV. Recurrent Nipah outbreaks in Kerala: implications for health policy and preparedness. Front Public Health. 2024;12:
1356515 . DOIPubMedGoogle Scholar
Figures
Cite This ArticleOriginal Publication Date: July 11, 2025
Table of Contents – Volume 31, Number 8—August 2025
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Emily S. Gurley, E6545, Johns Hopkins Bloomberg School of Public Health, 615 N Wolfe St, Baltimore, MD 21205, USA
Top