Volume 31, Number 9—September 2025
Research
Detection of Multiple Nosocomial Trichosporon asahii Transmission Events via Microsatellite Typing Assay, South America
Figure 2
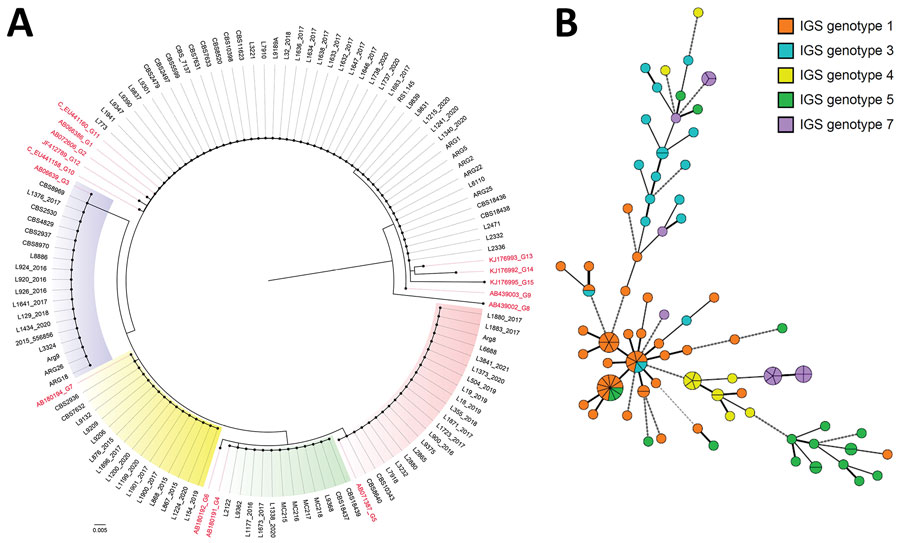
Figure 2. Phylogeny of multiple nosocomial Trichosporon asahii transmission events via microsatellite typing assay, South America. We compared IGS1 sequencing–based genotyping and a microsatellite panel for 111 T. asahii isolates. A) Neighbor-joining tree based on the rDNA sequencing target, conducted in MEGAX (https://www.megasoftware.net). Red text indicates GenBank accession numbers for reference strains of the described genotypes. Scale bar indicates nucleotide differences per site. B) Minimum spanning tree generated by 6 loci of microsatellite data showing the relationship between microsatellite typing and IGS1 genotypes. Each circle represents a unique genotype; if multiple isolates share an identical genotype, they are shown as a fraction of the circle. Lines between circles indicate the relative similarity, the shorter the line the less prominent the difference between the microsatellite genotypes. Thick lines identify genotypes with fewer differences (1 of 6 loci tested); medium thick lines identify genotypes with differences in 2 of the 6 loci; thick and medium thick dashed lines represent genotypes that share 2–3 identical loci of the 6 tested. Thin dotted lines identify genotypes sharing only 1 identical locus. IGS, intergenic spacer.