Volume 31, Number 9—September 2025
Dispatch
Gastric Submucosal Tumor in Patient Infected with Dioctophyme renale Roundworm, South Korea, 2024
Figure 3
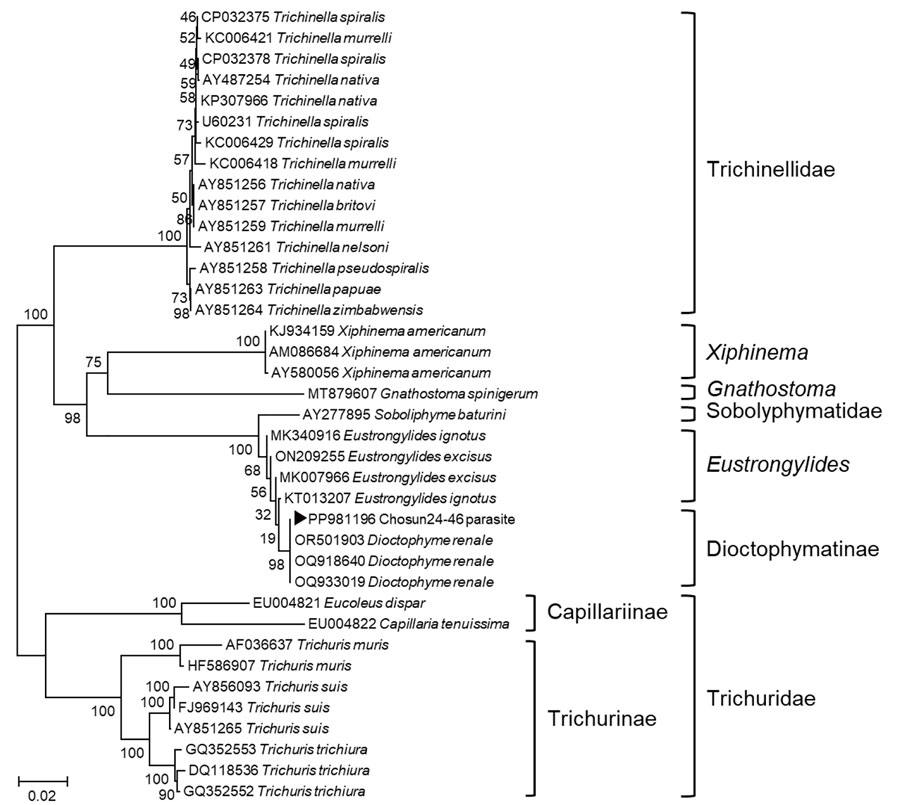
Figure 3. Phylogenetic tree for Dioctophyme renale roundworm showing a sequence from a patient with gastric submucosal tumor (black triangle), South Korea, 2024. Phylogenetic tree was based on the targeted 974-bp PCR amplicon sequence of D. renale from small subunit rRNA gene (18s RNA gene) sequences retrieved from GenBank (accession numbers shown). CLUSTAL X (http://www.clustal.org/clustal2) was used to construct phylogenetic tree by using neighbor-joining with 1,000 bootstrap replicates. We compared the D. renale sequence from our case with other roundworm sequences, and it aligned with 100% homology to D. renale. Scale bar indicates number of nucleotide substitutions per site.
1These authors contributed equally to this study.
Page created: May 22, 2025
Page updated: August 26, 2025
Page reviewed: August 26, 2025
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.