Volume 31, Number 9—September 2025
Dispatch
Novel Henipavirus, Salt Gully Virus, Isolated from Pteropid Bats, Australia
Abstract
We describe isolation and characterization of a novel henipavirus, designated Salt Gully virus, from the urine of pteropid bats in Australia. We noted the virus to be most closely related to Angavokely virus, not reliant on ephrin receptors for cell entry, and of unknown risk for human disease.
Bats of the genus Pteropus are natural reservoirs for zoonotic viruses, including the henipaviruses: enveloped, nonsegmented, negative-sense RNA viruses belonging to the family Paramyxoviridae (1). Hendra virus (HeV) and Nipah virus (NiV) represent the prototype henipaviruses and have caused fatal zoonotic spillover events from pteropid bats into animals and humans (2,3). High virulence, broad species tropism, and a lack of approved human vaccines and therapeutics classifies HeV and NiV as risk group 4 pathogens, restricting handling to Biosafety Level 4 (4).
Researchers first identified henipaviruses when an outbreak of HeV caused the death of 14 horses and 1 horse trainer in 1994 in Brisbane, Queensland, Australia (2). In total, 4 humans and >100 horses have died from HeV infection (5,6). In 2009, researchers isolated the first nonpathogenic henipavirus, Cedar virus (CedV), from pteropid bat urine collected during bat surveillance activities in Cedar Grove, Queensland, Australia (7).
Pteropid bats are the only natural reservoir identified within Australia for henipaviruses and, globally, detection of henipavirus relates most predominantly with pteropid bats. However, reports have noted an increasing number of novel henipa-like viruses detected in species of rodents, shrews, and opossums (8–10); such viruses have been classified by the International Committee on Taxonomy of Viruses in a new genus, Parahenipavirus (11).
We describe isolation and in vitro characterization of a novel pteropid bat–borne henipavirus in Australia. We obtained full-length sequences and assessed the virus’s genetic relationship to other henipaviruses. We also compared the virus’s growth, species tropism, and host cell receptor usage with HeV and CedV.
On July 11, 2011, we collected 30 pooled bat urine samples from a pteropid bat roost at Bicentennial Park, Boonah, Queensland, Australia, for an HeV surveillance project. We screened samples using quantitative real-time PCR for HeV. Ten samples were negative for HeV and inoculated onto Vero (African green monkey kidney) and primary Pteropus alecto kidney (PaKi) cell monolayers. We observed no viral cytopathic effects (CPE) after 7 days; however, the Vero tissue culture supernatant (TCSN) from 1 sample (BO13) tested positive when we employed generic reverse transcription PCR primers for paramyxovirus and henipavirus/morbillivirus (12). Sequencing of PCR products revealed a novel henipavirus sequence. Further passage of this virus in Vero cells yielded no CPE, and the virus did not replicate. However, when we inoculated TCSN from Vero cells onto PaKi cells, we observed replication and CPE. We then propagated a working stock in Vero cells, resulting in viral CPE demonstrating small syncytia, cell fusion, and rounded up cells (Appendix Figure). We designated the novel virus Salt Gully virus (SGV) based on the collection location.
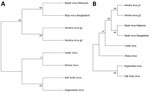
Next-generation sequencing of RNA extracted from TCSN on the Illumina platform (https://www.illumina.com), followed by genome assembly and analysis, revealed a large, complete genome of 19,884 nt (GenBank accession no. PV233879), adhering to the rule of 6 for paramyxoviruses (13). This genome included 6 distinct open reading frames that encoded 6 proteins: nucleocapsid (N), phosphoprotein (P), matrix (M), fusion (F), glycoprotein (G), and RNA polymerase (L). In addition, an alternative start codon within the P gene indicated the presence of a 7th open reading frame and a C protein, consistent with other henipavirus genomes. Whole-genome nucleotide alignment with other known henipaviruses showed that SGV shared 38% identity with HeV and NiV, 37% identity with Angavokely virus (AngV), and 35% identity with CedV and Ghana virus. We determined SGV to be phylogenetically most closely related to AngV (Figure 1).
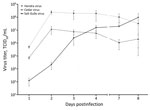
Investigating species tropism and growth kinetics, we found that SGV infected Vero, pteropid bat, and human cell lines, showing varying levels of CPE by 3 dpi. We observed no viral CPE in porcine or primary equine cell lines by 7 dpi. HeV replicated in all 5 cell lines, and CedV replicated in all except equine (Table). Initially, SGV displayed delayed replication in Vero cells; however, SGV reached maximum viral titer by 8 dpi at a titer comparable to HeV and CedV, which peaked by 2 dpi and then declined (Figure 2).
We used human epithelial (HeLa)-USU cells and recombinant HeLa-USU cell lines expressing ephrin-B2 or ephrin-B3 to assess SGV receptor usage. We observed viral CPE in HeLa cells expressing ephrin-B2 and ephrin-B3 for HeV and in ephrin-B2-expressing HeLa cells for CedV after 2 dpi. HeV and CedV did not infect HeLa-USU cells, as shown previously (14). In contrast, SGV caused CPE in all 3 HeLa cell lines after 5 dpi, indicating usage of an unknown receptor that is neither ephrin-B2 nor ephrin-B3 (Figure 3).
We detected SGV in bat urine samples collected in Australia in 2011 and successfully isolated the virus using pteropid bat cell lines. Initially, the CPE of inoculated Vero cells was minimal, requiring passage in pteropid kidney cells to achieve efficient virus replication. Despite the identification of multiple new henipa-like viruses in pteropid bats and small mammals globally, viral isolation is largely unsuccessful and remains a technical challenge. In our study, employing primary cell lines derived from the relevant species resulted in virus isolation.
Full-length genome sequencing of SGV revealed a genome organization consistent with other known henipaviruses, with all predicted henipavirus protein ORFs identified. Whole-genome alignments comparing SGV to other henipaviruses revealed 35%–38% identity at a nucleotide level. Of interest, phylogenetic analysis of the genome clusters SGV with AngV, a henipavirus that was detected in fruit bats (Eidolon dupreanum) in Madagascar in 2022 (15).
We assessed the ability for SGV to infect various mammalian cell lines in vitro, including Vero, PaKi, HeLa, equine fetal kidney, and porcine kidney cells. SGV’s notable ability to infect human cells underscores its potential for human infection. Unlike HeV, which infected all 5 cell lines, SGV did not cause CPE in porcine or equine cells. SGV could grow to high titer in Vero cells, albeit slower than HeV. Collectively, these results indicate SGV may not have the broad species tropism of pathogenic henipaviruses but could pose a human risk.
Ephrin-B2 and ephrin-B3 are host cell receptors for HeV and NiV (14), and the sequence conservation of those ligands across many species supports the broad species tropism of classical henipaviruses. The HeLa-USU cell line we used has been shown to lack expression of ephrin-B2 and ephrin-B3 and was used to determine the functional host cell receptor for CedV. In our study, SGV infected all 3 HeLa-USU cell lines, demonstrating that SGV cell entry is not reliant on ephrin-B2 or ephrin-B3, suggesting SGV uses a yet unidentified host cell receptor. In comparison, research has shown the glycoprotein G of Ghana virus could bind to ephrin-B2 (but not ephrin-B3), whereas predicted structure-based alignments suggest AngV is unlikely to use ephrin receptors for host cell entry (15). Further characterization is required to determine the functional host cell receptor for SGV and accurately assess pathogenicity in other species.
In summary, we identified, isolated, and characterized a novel henipavirus from pteropid bats in Australia. Amid the increasing discovery of novel henipa-like viruses in new locations and species, SGV is a true henipavirus and phylogenetically clusters with other bat henipaviruses. However, this virus’s pathogenicity remains unknown, making the susceptibility of human and animal populations in Australia uncertain.
Ms Barr is an experimental scientist at the CSIRO Australian Centre for Disease Preparedness in Geelong, Victoria, Australia. Her research interests include zoonotic batborne viruses, virus discovery, viruses requiring high-containment, and serologic assays.
Acknowledgment
We acknowledge Hume Field and team for assisting with urine collection from underneath pteropid bat colonies, James Gilkerson for providing the primary equine fetal kidney cell line used in this study, Christopher Broder for providing the HeLa-USU cell line used to study ephrin usage, and Kyle Catalan and Kim Blasdell for technical help with full-length sequencing.
References
- Rima B, Balkema-Buschmann A, Dundon WG, Duprex P, Easton A, Fouchier R, et al.; Ictv Report Consortium. ICTV virus taxonomy profile: Paramyxoviridae. J Gen Virol. 2019;100:1593–4. DOIPubMedGoogle Scholar
- Murray K, Selleck P, Hooper P, Hyatt A, Gould A, Gleeson L, et al. A morbillivirus that caused fatal disease in horses and humans. Science. 1995;268:94–7. DOIPubMedGoogle Scholar
- Chua KB, Bellini WJ, Rota PA, Harcourt BH, Tamin A, Lam SK, et al. Nipah virus: a recently emergent deadly paramyxovirus. Science. 2000;288:1432–5. DOIPubMedGoogle Scholar
- Middleton D, Pallister J, Klein R, Feng YR, Haining J, Arkinstall R, et al. Hendra virus vaccine, a one health approach to protecting horse, human, and environmental health. Emerg Infect Dis. 2014;20:372–9. DOIPubMedGoogle Scholar
- Queensland Government. Summary of Hendra virus incidents in horses. 2022 [cited 2025 Jul 23] https://www.business.qld.gov.au/industries/service-industries-professionals/service-industries/veterinary-surgeons/guidelines-hendra/incident-summary
- New South Wales Government. Summary of human cases of Hendra virus infection. 2022 [cited 2025 Jul 23] https://www.health.nsw.gov.au/Infectious/controlguideline/Pages/hendra-case-summary.aspx
- Marsh GA, de Jong C, Barr JA, Tachedjian M, Smith C, Middleton D, et al. Cedar virus: a novel Henipavirus isolated from Australian bats. PLoS Pathog. 2012;8:
e1002836 . DOIPubMedGoogle Scholar - Wu Z, Yang L, Yang F, Ren X, Jiang J, Dong J, et al. Novel henipa-like virus, Mojiang paramyxovirus, in rats, China, 2012. Emerg Infect Dis. 2014;20:1064–6. DOIPubMedGoogle Scholar
- Vanmechelen B, Meurs S, Horemans M, Loosen A, Joly Maes T, Laenen L, et al. The characterization of multiple novel paramyxoviruses highlights the diverse nature of the subfamily Orthoparamyxovirinae. Virus Evol. 2022;8:veac061. DOIGoogle Scholar
- Lee SH, Kim K, Kim J, No JS, Park K, Budhathoki S, et al. Discovery and genetic characterization of novel paramyxoviruses related to the genus Henipavirus in Crocidura species in the Republic of Korea. Viruses. 2021;13:2020. DOIPubMedGoogle Scholar
- Caruso S, Edwards SJ. Recently emerged novel Henipa-like viruses: shining a spotlight on the shrew. Viruses. 2023;15:2407. DOIPubMedGoogle Scholar
- Tong S, Chern SWW, Li Y, Pallansch MA, Anderson LJ. Sensitive and broadly reactive reverse transcription-PCR assays to detect novel paramyxoviruses. J Clin Microbiol. 2008;46:2652–8. DOIPubMedGoogle Scholar
- Kolakofsky D, Pelet T, Garcin D, Hausmann S, Curran J, Roux L. Paramyxovirus RNA synthesis and the requirement for hexamer genome length: the rule of six revisited. J Virol. 1998;72:891–9. DOIPubMedGoogle Scholar
- Bonaparte MI, Dimitrov AS, Bossart KN, Crameri G, Mungall BA, Bishop KA, et al. Ephrin-B2 ligand is a functional receptor for Hendra virus and Nipah virus. Proc Natl Acad Sci U S A. 2005;102:10652–7. DOIPubMedGoogle Scholar
- Madera S, Kistler A, Ranaivoson HC, Ahyong V, Andrianiaina A, Andry S, et al. Discovery and genomic characterization of a novel henipavirus, Angavokely virus, from fruit bats in Madagascar. J Virol. 2022;96:
e0092122 . DOIPubMedGoogle Scholar
Figures
Table
Cite This ArticleOriginal Publication Date: August 19, 2025
Table of Contents – Volume 31, Number 9—September 2025
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Jennifer Barr, CSIRO ACDP, 5 Portarlington Rd, East Geelong, VIC 3220, Australia
Top