Volume 31, Number 9—September 2025
Dispatch
Rapidly Progressing Melioidosis Outbreak in City Center Zoo, Hong Kong, 2024
Abstract
In October 2024, twelve primates from 4 species died of sepsis at the Hong Kong Zoological and Botanical Gardens. Postmortem examinations and microbiological analyses confirmed Burkholderia pseudomallei infection. Phylogenetic analysis revealed a clonal sequence type 46 strain with minimal variation, signifying a single source. This outbreak highlights melioidosis risk in zoo settings.
During October 2024, a series of sudden primate deaths occurred in the mammals section of the Hong Kong Zoological and Botanical Gardens (HKZBG), a popular city-center tourist attraction in Hong Kong, China, managed by the Leisure and Cultural Services Department. At the time of the outbreak, the facility housed 80 nonhuman primates (NHPs) across 12 species. We report the clinical course of illness and outcomes for the outbreak.
On the morning of October 13, 2024, four monkeys at HKZBG were found dead in their enclosures: 1 cotton-top tamarin (Saguinus oedipus), 2 white-faced sakis (Pithecia pithecia), and 1 common squirrel monkey (Saimiri sciureus). Concurrently, another 2 cotton-top tamarins, 1 white-faced saki, and 1 De Brazza’s monkey (Cercopithecus neglectus) with clinical signs of varying severity (e.g., lethargy, anorexia, fever) were sent to the zoo clinic for examination and emergency care; all 4 monkeys died later that day. Upon observed clinical signs, 1 additional white-faced saki and 1 De Brazza’s monkey were isolated for intensive monitoring; the saki died on October 14 and the De Brazza’s monkey on October 22. Another 2 common squirrel monkeys hospitalized on October 16 died on October 19 and October 20 (Table; Appendix Figure 1).
In total, 12 monkeys spanning 4 species eventually died in this outbreak. Species-specific mortality rates were high: 2 (50%) of 4 De Brazza’s monkeys, 3 (75%) of 4 common squirrel monkeys, 3 (50%) of 6 cotton-top tamarins, and 4 (36.4%) of 11 white-faced saki died. Collectively, the deaths represented 15% of the zoo’s NHP population and a 48% mortality rate among the 25 animals comprising the 4 species, underscoring the outbreak’s severity.
To safeguard public and animal health, the mammals section of the zoo was temporarily closed starting on October 14. All enclosures were thoroughly cleaned and disinfected. The remaining animals in the section were clinically healthy. Health monitoring for staff who take care of animals was provided, and health conditions of staff were unremarkable.
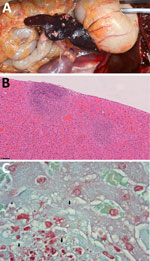
A government working group conducted comprehensive follow-up actions, including postmortem examinations and diagnostic testing, to investigate the monkey deaths. On October 18, laboratory results confirmed that all tested monkeys had died from sepsis caused by Burkholderia pseudomallei infection. The HKZBG veterinarian performed postmortem examination and tissue sampling of the 8 monkeys that initially died on October 13. The remaining 4 monkeys were sent to the Tai Lung Veterinary Laboratory under the Agriculture, Fisheries and Conservation Department of the Government of the Hong Kong Special Administrative Region for postmortem examination, histopathology, and microbiological testing. Gross and histopathologic findings of all animals demonstrated that the liver and spleen were the most severely affected organs, characterized by acute necrotizing to necrosuppurative splenitis (Figure 1, panel A) and hepatitis (Figure 1, panel B); intralesional gram-negative bacilli were detected (Figure 1, panel C). Evidence of hematogenous spread to the lungs was also present in some monkeys, resulting in mild fibrinonecrotic interstitial pneumonia.
Bacterial cultures yielded 11 pure isolates, which were initially identified as Burkholderia spp. using a Biotyper matrix-assisted laser desorption/ionization time-of-flight mass spectrometry system (Bruker, https://www.bruker.com). Real-time PCR targeting a 115-bp fragment of the type III secretion system confirmed B. pseudomallei (1). Additional molecular screenings for monkeypox virus, coronavirus, SARS-CoV-2, Leptospira spp., and influenza A virus all produced negative results.
Initial investigations hypothesized an environmental source, including possible release from recent soil disturbances in early October. A comprehensive environmental assessment was conducted, including PCR and culture of 25 soil, 27 drinking water, 10 feed supplement, and 8 environmental samples. All samples tested negative for B. pseudomallei (Appendix).
B. pseudomallei was cultured from the liver, lungs, or spleen of 11 of the 12 dead animals. The isolates underwent whole-genome sequencing (Appendix), and a hybrid assembly approach using Hybracter version 0.11.0 (https://github.com/gbouras13/hybracter) generated high-quality, closed-gap complete genomes (2). Multilocus sequence typing (MLST) based on profiles from the PubMLST database (https://pubmlst.org) classified all isolates as sequence type (ST) 46 and core-genome MLST (cgMLST) type 1070 (3).
For phylogenetic context, we constructed a maximum-likelihood phylogeny comparing study isolates to 40 reference B. pseudomallei genomes representing ST46 and closely related sequence types from GenBank (4). The tree used single-nucleotide polymorphisms (SNPs) derived from 3,127 single-copy cgMLST genes (5) (Appendix). Results showed that the 11 Hong Kong isolates clustered together in a monophyletic clade with 100% bootstrap confidence (Appendix Figure 2), exhibiting exceptionally tight genetic relatedness of only 0–1 core-genome SNP (cgSNP) differences. In addition to core-genome phylogeny, whole-genome average nucleotide identity analysis further confirmed high genetic similarity (99.99839%–99.99954%) among the 11 isolates (Appendix Figure 3). Considering that B. pseudomallei can develop 8 SNPs during a 12-day acute infection period (6), the minimal SNP variation among the HKZBG isolates suggests a single clonal infection source (7), likely from a singular introduction event rather than sustained transmission among the animals.
Focused comparative analysis of ST46 (Figure 2) revealed that the HKZBG clade was most closely related to a clonal cluster of 3 strains from northern Hainan Province, China, with a genetic distance of 18 cgSNPs. The next closest relatives were strains from Australia (27 cgSNP difference) and Thailand (31 cgSNP difference). Such minimal divergence underscores that ST46 is a recurring sequence type within the Asia–Oceania region. Although that ST is the third most frequently reported in the global PubMLST database and has been isolated from humans, the environment, and other animals, including monkeys (8,9), it had not been previously reported in Hong Kong. Moreover, the isolates from this outbreak are genetically distinct from the local outbreak strain ST1996 reported in 2022 (10) and from other local sequence types, such as ST70, ST37, and ST32 (11) (Figure 3). Among globally reported ST46 strains, the closest relatives to the HKZBG clade were strains isolated in 2002–2003 from northern Hainan Province, China (National Center for Biotechnology Information Assembly database accession nos. GCA_015312861.1, GCA_015312871.1, and GCA_015312851.1) (Figure 2). Those isolates share the same cgMLST type 1070 profile, suggesting that this lineage has been established in southern China for decades (12).
The rapid and nearly simultaneous deaths of multiple primates, together with the swift progression of the disease, suggest that this melioidosis outbreak was the result of a concentrated or highly virulent exposure event (13). Postmortem findings revealed extensive hepatic and splenic involvement in all animals. Although some animals exhibited pneumonia, pulmonary lesions were mild, and the pattern was characteristic of hematogenous spread to the lungs rather than bronchogenic. That finding is different from the lesions observed in NHP models after aerogenous infection (14,15).
Despite extensive investigations, including environmental sampling and genomic analysis, the precise source of infection in this outbreak remains unidentified. The initial hypothesis that soil disturbances released environmental B. pseudomallei was not supported because of negative environmental results and because most affected monkeys had long-term residency at the zoo (many >6 years) with no history of melioidosis.
In reaction to this incident, the zoo implemented stringent biosecurity measures, including thorough enclosure disinfection and restricted access to affected areas. No further monkey deaths were recorded after October 22, and no cases of human melioidosis were noted during the investigation period.
This outbreak, which resulted in the loss of 12 monkeys, including critically endangered cotton-top tamarins, highlights the potential threat of melioidosis in zoologic settings. Climate change potentially could increase the incidence of B. pseudomallei infections, even in urban environments like Hong Kong, requiring enhanced biosecurity, vigilant health monitoring, and a high index of suspicion for melioidosis in cases of unusual illnesses and deaths in captive wildlife. Such proactive measures are critical for protecting both animal and human health.
Dr. Brackman is a senior veterinary officer in the Agriculture, Fisheries and Conservation Department, Government of the Hong Kong Special Administrative Region, China. His research interests are focused on the field of One Health, in particular detection and characterization of zoonotic and transboundary diseases, including avian influenza and African swine fever, across wildlife and livestock.
Acknowledgments
We thank the staff of the Tai Lung Veterinary Laboratory for their technical expertise in isolating and identifying the clinical strains of B. pseudomallei. We are particularly indebted to the HKZBG team for their prompt implementation of stringent biosecurity measures following the initial primate mortality report and for their thorough investigation that contributed significantly to understanding the outbreak.
The whole-genome sequencing data generated for this study have been deposited in the National Center for Biotechnology Information Sequence Read Archive (BioProject accession no. PRJNA1271261; individual accession nos. SRR33822591–612).
References
- Novak RT, Glass MB, Gee JE, Gal D, Mayo MJ, Currie BJ, et al. Development and evaluation of a real-time PCR assay targeting the type III secretion system of Burkholderia pseudomallei. J Clin Microbiol. 2006;44:85–90. DOIPubMedGoogle Scholar
- Bouras G, Houtak G, Wick RR, Mallawaarachchi V, Roach MJ, Papudeshi B, et al. Hybracter: enabling scalable, automated, complete and accurate bacterial genome assemblies. Microb Genom. 2024;10:
001244 . DOIPubMedGoogle Scholar - Jolley KA, Bray JE, Maiden MCJ. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018;3:124. DOIPubMedGoogle Scholar
- Fang Y, Hu Z, Chen H, Gu J, Hu H, Qu L, et al. Multilocus sequencing-based evolutionary analysis of 52 strains of Burkholderia pseudomallei in Hainan, China. Epidemiol Infect. 2018;147:
e22 . DOIPubMedGoogle Scholar - Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–74. DOIPubMedGoogle Scholar
- Limmathurotsakul D, Holden MT, Coupland P, Price EP, Chantratita N, Wuthiekanun V, et al. Microevolution of Burkholderia pseudomallei during an acute infection. J Clin Microbiol. 2014;52:3418–21. DOIPubMedGoogle Scholar
- Meumann EM, Kaestli M, Mayo M, Ward L, Rachlin A, Webb JR, et al. Emergence of Burkholderia pseudomallei sequence type 562, northern Australia. Emerg Infect Dis. 2021;27:1057–67. DOIPubMedGoogle Scholar
- Godoy D, Randle G, Simpson AJ, Aanensen DM, Pitt TL, Kinoshita R, et al. Multilocus sequence typing and evolutionary relationships among the causative agents of melioidosis and glanders, Burkholderia pseudomallei and Burkholderia mallei. J Clin Microbiol. 2003;41:2068–79. DOIPubMedGoogle Scholar
- Zhu X, Chen H, Li S, Wang LC, Wu DR, Wang XM, et al. Molecular characteristics of Burkholderia pseudomallei collected from humans in Hainan, China. Front Microbiol. 2020;11:778. DOIPubMedGoogle Scholar
- Wu WG, Shum MH, Wong IT, Lu KK, Lee LK, Leung JS, et al. Probable airborne transmission of Burkholderia pseudomallei causing an urban outbreak of melioidosis during typhoon season in Hong Kong, China. Emerg Microbes Infect. 2023;12:
2204155 . DOIPubMedGoogle Scholar - Lui G, Tam A, Tso EYK, Wu AKL, Zee J, Choi KW, et al. Melioidosis in Hong Kong. Trop Med Infect Dis. 2018;3:91. DOIPubMedGoogle Scholar
- Zheng H, Qin J, Chen H, Hu H, Zhang X, Yang C, et al. Genetic diversity and transmission patterns of Burkholderia pseudomallei on Hainan island, China, revealed by a population genomics analysis. Microb Genom. 2021;7:
000659 . DOIPubMedGoogle Scholar - Nelson M, Nunez A, Ngugi SA, Atkins TP. The lymphatic system as a potential mechanism of spread of melioidosis following ingestion of Burkholderia pseudomallei. PLoS Negl Trop Dis. 2021;15:
e0009016 . DOIPubMedGoogle Scholar - Nelson M, Nunez A, Ngugi SA, Sinclair A, Atkins TP. Characterization of lesion formation in marmosets following inhalational challenge with different strains of Burkholderia pseudomallei. Int J Exp Pathol. 2015;96:414–26. DOIPubMedGoogle Scholar
- Trevino SR, Dankmeyer JL, Fetterer DP, Klimko CP, Raymond JLW, Moreau AM, et al. Comparative virulence of three different strains of Burkholderia pseudomallei in an aerosol non-human primate model. PLoS Negl Trop Dis. 2021;15:
e0009125 . DOIPubMedGoogle Scholar
Figures
Table
Cite This ArticleOriginal Publication Date: August 11, 2025
1These authors contributed equally to this article.
Table of Contents – Volume 31, Number 9—September 2025
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Franklin Wang-Ngai Chow or Gilman Kit-Hang Sui, Hong Kong Polytechnic University, Health Technology and Informatics, Y928, 9/F, Block Y, Kowloon, Hong Kong 999077, China
Top