Volume 7, Number 5—October 2001
Research
Clonal Expansion of Sequence Type (ST-)5 and Emergence of ST-7 in Serogroup A Meningococci, Africa
Cite This Article
Citation for Media
Abstract
One hundred four serogroup A meningococci in our collection, isolated in Africa from 1988 to 1999, were characterized by multilocus sequence typing (MLST). Our results and data from the Internet indicate that sequence type 5 (ST-5) strains were responsible for most of African outbreaks and sporadic cases during this period. In 1995, a new clone, characterized by ST-7 sequence, emerged and was responsible for severe outbreaks in Chad (1998) and Sudan (1999). MLST and epidemiologic data indicate that ST-5 and ST-7 represent two virulent clones. These two STs, which belong to subgroup III, differ only in the pgm locus: allele pgm3 is characteristic for ST-5 and allele pgm19 for ST-7. Subgroup III strains were responsible for two pandemics in the 1960s and 1980s. Our data show that the third subgroup III pandemic has now reached Africa.
Multilocus enzyme electrophoresis (MLEE) has been the reference method for global epidemiology of Neisseria meningitidis. This method identified clusters of closely related strains (for example, subgroup III for serogroup A N. meningitidis and ET-5 complex for serogroup B N. meningitidis) and permitted monitoring of their clonal spread throughout the world (1-3). This method, however, relies on the indirect assignment of alleles based on the electrophoretic mobility of enzymes. However, indistinguishable variants may be encoded by very different sequences, and results obtained in different laboratories may be difficult to compare. Rather than comparing the electrophoretic mobilities of the enzymes they encode, Maiden et al. adapted this method by identifying alleles directly from the nucleotide sequences of internal fragments of housekeeping genes (4). This new method, called multilocus sequence typing (MLST), is based on the sequencing of DNA fragments belonging to seven housekeeping genes. MLST results are unambiguous and distinguish more alleles per locus, allowing high-level discriminations between isolates. The first data published by Maiden showed good congruence between MLST and MLEE (4).
The aim of our study was to check MLST for the characterization of 104 serogroup A N. meningitidis in our collection, isolated in 14 African countries from 1988 to 1999, to determine the feasibility of the technique and which sequence types were circulating in some African countries during this period.
Bacterial Strains
A total of 104 serogroup A N. meningitidis strains isolated in 14 African countries from 1988 to 1999 and received at the World Health Organization Collaborating Centre in Marseilles were included in this study; 101 were isolated from cerebrospinal fluid (CSF), 1 from blood culture of a patient with meningococcal meningitis, and 2 from pharynx (Table). For some outbreaks, we randomly chose three strains for sequencing, if the pulsed-field gel electrophoresis (PFGE) fingerprint patterns were identical (Chad 1988, Central African Republic 1992, and Senegal 1998 outbreaks). Since 1999, all meningococcal strains have been routinely assayed by MLST.
All these strains were stored at -80°C in brain heart broth with 15% glycerol. The identification number for each strain was preceded by Mrs for Marseilles.
Bacterial Identification, Serogrouping, Typing, Subtyping
Bacterial identification was carried out by Gram staining, oxidase test, and tests for biochemical characteristics by using a ready-for-use kit (Neisseria 4H Sanofi Pasteur, Paris, France). N. meningitidis strains were serogrouped by agglutination with sera manufactured in Institut de Médecine Tropicale du Service de Santé des Armées (Marseilles). Serotypes and subtypes were determined by using the monoclonal kit from the National Institute of Public Health and the Environment (Bilthoven, the Netherlands) and the whole-cell enzyme immunoassay technique described elsewhere (5,6; Abdillahi, unpub. data).
Whole chromosomal DNA was compared by PFGE of macrorestriction fragments generated by endonuclease Bgl II (7). Agar plugs containing bacteria were treated by lysozyme, Proteinase K, and then Pefabloc (Roche, Meylan, France). Plugs were incubated with 25 U of the endonuclease Bgl II (Eurogentec, Seraing, Belgium) overnight at 37°C. Electrophoresis was performed with a CHEF Mapper (Bio-Rad Laboratories, Hercules, CA) in 0.5x Tris-borate-EDTA at 14°C, and a voltage of 4.5 V/cm was applied with a pulse-time ramping from 30 seconds to 1 second over 22 hr. Then a pulse of 0.1 to 1 second was applied for 2 hours and 30 minutes with a voltage of 6 V/cm. Fingerprint patterns were analyzed by using Tenover criteria (8).
For MLST, the primers of the housekeeping genes abcZ (putative ABC transporter), adk (adenylate kinase), aroE (shikimate dehydrogenase), gdh (glucose-6-phosphate dehydrogenase), pdhC (pyruvate dehydrogenase subunit), and pgm (phosphoglucomutase) were synthesized by our institute according to the sequences published by Maiden et al. (4). A seventh locus, fumC (fumarase), was added. Primers for amplification and sequencing of fumC fragment were synthesized from the sequences given on the MLST web site, http://www.mlst.net. After DNA preparation and amplification by polymerase chain reaction (PCR), each locus sequence was analyzed on an ABI prism 310 Genetic Analyzer (Perkin-Elmer [PE] Applied Biosystems, Foster City, CA) or ABI prism 373 DNA Sequencer (PE Applied Biosystems). The sequence alignment was performed on the Sequence Navigator software (PE Applied Biosystems). The sequences were then compared with the different existing alleles registered on the MLST web site.
Bacterial Identification, Grouping, Typing, Subtyping
All 104 strains were gram-negative diplococci, oxidase positive, and catalase positive. They were classified as N. meningitidis on the basis of growth characteristics on selective medium, acidification of glucose and maltose, and gamma-glutamyl transferase activity (9). One hundred three strains were serogroup A, type 4, and subtype P1.9 (A:4:P1.9), the same formula as strains belonging to subgroup III. One strain isolated in Algeria in 1992 was A:4:P1.10.
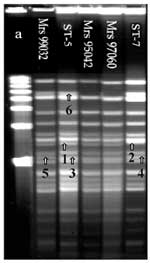
DNA fingerprint patterns generated with endonuclease Bgl II and analyzed by PFGE showed 103 closely related fingerprint patterns. Two profiles could be identified. ST-5 was the first pattern found in Africa and the most frequently isolated from 1988 to 1996 (7). The second profile was the ST-7 pattern, attributable to strains isolated more recently in Algeria, Cameroon, Sudan, Chad, and Niger. The two patterns are closely related but have four band differences (Figure). Most strains analyzed by PFGE were indistinguishable from ST-5 or ST-7. However, one strain (Mrs 99032) isolated in Dakar (1999) from CSF, showed two band differences with the outbreak pattern; and two strains (Mrs 95042, Mrs 97060), isolated in Burkina Faso in 1995 and 1997, showed one band difference with the ST-5 pattern. Strains with the ST-7 profile had almost the same patterns, but one strain (Mrs 98118), isolated in Zaire in 1998, showed one band difference. One serogroup A:4:P1.10 meningococcus (Mrs 92060), isolated in 1992 in Algeria, had a totally different profile; this strain was not related to the other 103 strains. All strains isolated from outbreaks in Chad (1988), Central African Republic (1992), and Senegal (1998) had identical PFGE fingerprint patterns; we sequenced three strains randomly chosen from these isolates (data not shown).
The MLST comparison between sequences and existing alleles allowed us to assign our sequences allele numbers (Table). On the MLST web site, the allele combination assigned 100 of 104 strains to ST-5 or ST-7 because they were identical to the consensus at seven loci. One strain (Mrs 92060), isolated in Algeria in 1992, was assigned to ST-1. Three strains had new STs: two strains (Mrs 95042, Mrs 97060), isolated in Burkina Faso in 1995 and 1997, were abcZ2 and assigned to ST-580, and one strain (Mrs 99066), isolated during the 1999 Senegal outbreak, was adk64 and assigned to ST-581.
MLST technique was established in our laboratory at the beginning of 1999. Since 2000, all meningococcal strains we have received have been routinely characterized by this method. The seven loci of the 104 serogroup A N. meningitidis included in this study were characterized by their sequences, and alleles were assigned directly at the MLST web site (http://www.mlst.net), resulting in identification of sequence types ST-1, ST-5, or ST-7 (Table). An e-mail with sequence trace files was sent to the MLST web site to obtain alleles and sequence types of three strains, subsequently classified as ST-580 or ST-581 (Table). Except for ST-1, identified in strain Mrs 92061, ST-7, ST-580 and ST-581 differ from ST-5 in only one locus, and these four related STs belong to subgroup III. One hundred of 104 strains were either ST-5 or ST-7. These two STs are closely related, differing only in pgm locus: pgm3 is characteristic for ST-5 and pgm19 for ST-7. These two alleles differ in sequence of 47 of their base pairs, most likely because of recombination, frequently seen in N. meningitidis (10,11). The surface epitopes of strains analyzed in this study (e.g, serogroup, serotype, and serosubtype) are identical in the strains of these two STs. However, a four-band difference was observed in the PFGE patterns, even though there is no BglII restriction site in either locus pgm3 or locus pgm19.
MLST and epidemiologic data indicate that strains of ST-5 and ST-7 represent two virulent clones (Table). To date, ST-5 strains have been isolated from several outbreaks: Chad (1988), Central African Republic and Burundi (1992), Cameroon (1993), Niger (1995, 1996), Burkina Faso (1996), Mali (1997), Senegal (1998 and 1999), and Guinea Bissau (1999) (Table). ST-5 strains were also isolated from a carrier returning from Saudi Arabia in 1987 (Centers for Disease Control and Prevention, unpub. data), Gambia in 1997 (Greenwood, unpub. data), and Ghana in 1997 and 1998 (Popovic, Pluschke, unpub. data). In 1995, when ST-5 strains were widespread, a new clone, characterized by ST-7, emerged. The oldest ST-7 in our collection is from Algeria (1995). ST-7 strains were isolated throughout the "meningitis belt" (12): Chad (1997, 1998, and 1999), Cameroon (1997, 1998, and 1999), Zaire (1998), Niger (1999), and Sudan (1999) (Table).
During the 11-year period 1988-1999, the ST-5 epidemic wave reached all African countries in the meningitis belt. In 1995, the ST-7 clone appeared in Algeria. The origin and emergence of the ST-7 clone might be explained by recombination events' conferring selective advantages to ST-7. Another possibility is that the homogenizing effect of a sequential bottleneck might have selected at random a ST-7 clone among a limited number of different genotypes, resulting in a population uniform for the new variant (13,14). Since their appearance in 1997, only strains of ST-7 have been identified in Chad and Cameroon. Similarly, the 1999 Sudan outbreak was due to an ST-7 clone. Therefore, it appears that ST-7 is a clonal replacement for ST-5 in African countries. Prospective monitoring and analysis of the isolates by MLST will be crucial for assessing the full significance of our observations.
Serogroup A strains of subgroup III were associated with the first pandemic that started in China in the mid-1960s and subsequently spread to Russia, Scandinavia, and Brazil. In the early 1980s, a second wave of meningococcal disease caused by subgroup III clones began in China, spread through Nepal and probably India, and then reached Saudi Arabia in 1987 (2,3,15, 16). Given that this particular clonal group had not been isolated in Africa before 1987, we speculate that subgroup III, and more precisely the ST-5 clone, was introduced into Africa in 1987 by pilgrims returning from Mecca (17-19). By 1988, epidemics of meningococcal disease were recorded in Chad and Sudan, eventually reaching most African countries (20; MLST web site). In 1995 and 1997, the ST-7 clone emerged in Africa and appears to be responsible for a new wave of epidemics.
Achtman et al. showed that the third pandemic caused by subgroup III began in China in 1993, causing large epidemics in Mongolia in 1994 and Moscow in 1996 (21). The strain associated with this pandemic can be readily recognized by the presence of the pgm19 allele. Results of our study suggest that this third pandemic has now reached Africa.
Implications of the ST-7 clone's replacing ST-5 can be substantial. Chad and probably Sudan experienced epidemics caused by ST-5 in 1988. Although ST-5 and ST-7 are closely related clones, herd immunity due to the presence of the ST-5 strains is now apparently surpassed since these two countries had severe outbreaks caused by the ST-7 clone in 1998 and 1999.
Justified concern is raised now that the new pandemic of subgroup III will spread to other countries of the meningitis belt. It is important to alert those countries, particularly Cameroon and Niger, where sporadic ST-7 strains are already present.
Among our 104 strains, 4 did not belong to ST-5 or ST-7 and were characterized by sequences ST-1, ST-580, and ST-581. The ST-1 strain, belonging to subgroup I, was isolated in Algeria in 1992. Subgroup I has been responsible for epidemics and sporadic cases in Africa since 1961. Although it has not been isolated for many years in the meningitis belt, it was still the predominant clone in South Africa in 1996 (2,3,22). In addition to three strains of ST-5 isolated in Burkina Faso in 1995 and 1996, two strains isolated in 1995 and 1997 were ST-580. This particular type is closely related to ST-5, differing only at the abcZ locus. Isolation of only a couple of strains of ST-580 substantially hampers speculation that ST-580 is a genetic variant that could potentially emerge. Finally, the third ST that differed from ST-5 was ST-581, which had a new adk allele (Table). A strain of this type was isolated from the CSF of a patient in Senegal in 1999. However, this new clone will probably not emerge as a virulent clone (i.e., to replace ST-5); indeed, it will probably be lost in the future.
Although MLST makes standardization and interlaboratory comparison easier, the technique is time-consuming and expensive. Some simplifications may be possible. For example, 11 strains isolated during the 1999 Sudan outbreak showed the same PFGE fingerprint patterns as well as identical ST. In this case, sequencing only one strain would have been sufficient. However, strain Mrs 99066 of ST-581, isolated in Dakar in 1999, had a PFGE pattern identical to that typically seen in the strains of the ST-5 pattern. With few such exceptions, Bgl II PFGE analysis resulted in easy differentiation of ST-5 and ST-7 strains. Their PFGE patterns remained unchanged over several years and in strains isolated in different countries. Although it is accepted that PFGE is not appropriate for long-term comparison purposes, it may discriminate ST-5 from ST-7 strains of N. meningitidis serogroup A. Also, in epidemiologic investigations, differentiation of pgm3 from pgm19 in serogroup A strains recently isolated in Africa may be useful for differentiating ST-5 and ST-7. That could be accomplished by sequencing or by RFLP-PCR of pgm locus. However, the disadvantage of this approach is that strains of ST-580 would be identified as belonging to ST-5.
Since we established MLST in our laboratory in 1999, it has allowed us to obtain reliable and portable data that could be easily compared between laboratories without having to exchange strains. MLST was initially developed for studies of meningococcal population genetics. In this study, MLST was used along with epidemiologic data to identify two virulent clones characterized by ST-5 and ST-7. Strains of the ST-5 were responsible for the second pandemic wave that started in Africa in 1988, and the appearance of ST-7 strains may likely be responsible for the third one. In our study, MLST has proven to be a reliable and useful tool for molecular typing of N. meningitidis serogroup A and could replace MLEE as the standard for molecular typing. MLST may aid in identifying and monitoring the global spread of virulent STs to allow rapid implementation of preventive measures.
Dr. Nicolas is a physician, head of Unité du Méningocoque, World Health Organization (WHO) Collaborating Center in Marseille, France. The laboratory is a reference laboratory on meningococci for French Armed Forces, WHO, and African laboratories. Dr. Nicolas' research interests focus on population genetics and molecular epidemiology of Neisseria meningitidis.
Acknowledgments
The authors thank the following for bacterial strains: J.M Alonso; J.P. Chippaux; S. Djibo; I. Lisse; P. Colbachini; B. N'Doye; G. Raphenon; J.P. Boyer; P. Martin; J. Ahi Koffi; F. Coulom Pontier; E. Tikhomirov; Nageeb Sulaiman Saeed; biologists from Institut National de Recherche en Santé Publique, Bamako (Mali); physicians from the French Military Bioforce; H. Tali-Maamar; and J.B. Ndihokubwayo. We also thank M. Torrentino and B. Pastorino for sequencing; H. Pugelli for primer synthesis; M. Achtman and H. Tolou for comments on the manuscript; T. Popovic for help in preparing the manuscript; and E. Tikhomirov and D. Schaaf for epidemiologic data.
This work was supported in part by funds from Ministère de la Défense (France) (DGA/PEA 98 08 14, contract 98 100 60) and by funds from the World Health Organization (C11/181/2 (A). This publication made use of the MLST web site http://www.mlst.net, developed by Man-Suen Chan and situated at the University of Oxford. The development of this site is funded by the Wellcome Trust.
References
- Selander RK, Caugant DA, Ochman H, Muser JM, Gilmour MN, Whittman TS. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl Environ Microbiol. 1986;51:873–84.PubMedGoogle Scholar
- Caugant DA. Population genetics and molecular epidemiology of Neisseria meningitidis. APMIS. 1998;106:505–25. DOIPubMedGoogle Scholar
- Wang J-F, Caugant DA, Li X, Hu X, Poolman JT, Crowe BA, Clonal and antigenic analysis of serogroup A Neisseria meningitidis with particular reference to epidemiological features of epidemic meningitidis in the People's Republic of China. Infect Immun. 1992;60:5267–82.PubMedGoogle Scholar
- Maiden MCJ, Bygraves JA, Feil E, Morelli G, Russel J, Urwin R, Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci U S A. 1998;95:3140–5. DOIPubMedGoogle Scholar
- Frasch CE, Zollinger WD, Poolman JT. Serotype antigens of Neisseria meningitidis and a proposed scheme for designation of serotypes. Rev Infect Dis. 1985;7:504–10. DOIPubMedGoogle Scholar
- Poolman JT, Abdillahi H. Outer membrane protein serosubtyping of Neisseria meningitidis. Eur J Clin Microbiol Infect Dis. 1988;7:291–3. DOIPubMedGoogle Scholar
- Nicolas P, Parzy D, Martet G. Pulsed-field gel electrophoresis of clonal relationships among Neisseria meningitidis strains from different outbreaks. Eur J Clin Microbiol Infect Dis. 1997;16:541–4. DOIPubMedGoogle Scholar
- Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–9.PubMedGoogle Scholar
- Riou JY, Guibourdenche M. Méthodes de laboratoire Neisseria et Branhamella ISBN 2-901 320-09-0 Ed. Paris: Institut Pasteur; 1993.
- Feil EJ, Maiden MCJ, Achtman M, Spratt BG. The relative contribution of recombination and mutation to the divergence of clones of Neisseria meningitidis. Mol Biol Evol. 1999;16:1496–502.PubMedGoogle Scholar
- Maiden MCJ, Malorny B, Achtman M. A global gene pool in the Neisseriae. Mol Microbiol. 1996;21:1297–8. DOIPubMedGoogle Scholar
- Lapeyssonnie L. La méningite cérébrospinale en Afrique. Bull World Health Organ. 1963;28(Suppl):1–100.PubMedGoogle Scholar
- Morelli G, Malorny B, Müller K, Seiler A, Wang J-F, del Valle J, Clonal descent and microevolution of Neisseria meningitidis during 30 years of epidemic spread. Mol Microbiol. 1997;25:1047–64. DOIPubMedGoogle Scholar
- Achtman M. Microevolution and epidemic spread of serogroup A Neisseria meningitidis, a review. Gene. 1997;192:135–40. DOIPubMedGoogle Scholar
- Moore PS, Reeves MW, Schwarz B, Gellin BG, Broome CV. Intercontinental spread of an epidemic group A Neisseria meningitidis strain. Lancet. 1989;ii:260–3. DOIPubMedGoogle Scholar
- Achtman M, Kusecek B, Morelli G, Eickmann K, Wang J-F, Crowe B, A comparison of the variable antigens expressed by clone IV-1 and subgroup III of Neisseria meningitidis serogroup A. J Infect Dis. 1992;165:53–68. DOIPubMedGoogle Scholar
- Ministry of Health Annual Health Report. Saudi Arabia: The Ministry; 1987 (1407 Hijra). p. 279.
- Wahdan MH. Epidemiology of meningococcal meningitis: an overview of the situation in the eastern Mediterranean region. Intercountry meeting on preparedness and response to meningococcal meningitis outbreaks. Damascus. (WHO:EM/INC. MTG. PPD. REPNMO/4). Geneva: World Health Organization; 1989.
- Moore PS, Harrison LH, Telzak EE, Ajello GW, Broome CV. Group A meningococcal carriage in travelers returning from Saudi Arabia. JAMA. 1988;260:2686–9. DOIPubMedGoogle Scholar
- Nicolas P, Raphenon G, Guibourdenche M, Decousset L, Stor R, Gaye AB. The 1998 Senegal epidemic of meningitis was due to the clonal expansion of A:4:P1.9, clone III-1, sequence type 5 Neisseria meningitidis strains. J Clin Microbiol. 2000;38:198–200.PubMedGoogle Scholar
- Achtman M, van der Ende A, Zhu P, Koroleva IS, Kusecek B, Morelli G, Molecular epidemiology of four successive waves of serogroup A meningococcal disease in Moscow, Russia between 1969 and 1997. Emerg Infect Dis. 2001;7:420–7.PubMedGoogle Scholar
- Olyhoeck T, Crowe BA, Achtman M. Clonal population structure of Neisseria meningitidis serogroup A isolates from epidemics and pandemics between 1915 and 1983. Rev Infect Dis. 1987;9:665–92. DOIPubMedGoogle Scholar
Figure
Table
Cite This ArticleTable of Contents – Volume 7, Number 5—October 2001
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Pierre Nicolas, Unité du Méningocoque, IMTSSA, WHO Collaborating Center, BP 46, le Pharo, 13998 Marseille Armées, France; fax: 33 4 91 59 44 77
Top