Volume 8, Number 7—July 2002
Research
Infection by Ralstonia Species in Cystic Fibrosis Patients: Identification of R. pickettii and R. mannitolilytica by Polymerase Chain Reaction
Cite This Article
Citation for Media
Abstract
The frequency of respiratory tract infections caused by Ralstonia species in persons with cystic fibrosis (CF) and the role of these species in CF pulmonary disease are not well documented. In part, this lack of documentation may be attributed to the difficulty in accurately identifying Ralstonia species; R. mannitolilytica and R. pickettii in particular may be misidentified as other closely related species, particularly those of the Burkholderia cepacia complex. We used polyphasic analyses to identify 42 Ralstonia isolates from sputum cultures from 38 CF patients. Several isolates that could not be identified to the species level may belong to novel Ralstonia species. We demonstrated chronic colonization by using genotyping of serial isolates recovered from the same patient. To facilitate identification of R. mannitolilytica and R. pickettii, we developed 16S ribosomal DNA-based polymerase chain reaction assays that allow sensitive and specific identification of these species.
Cystic fibrosis (CF) is the most frequent hereditary disease in Caucasian populations (1); chronic microbial colonization of the large airways, leading to exacerbations of pulmonary infection, is the major cause of illness and death in CF patients. Typical CF pathogens include Staphylococcus aureus, Pseudomonas aeruginosa, Haemophilus influenzae, and Burkholderia cepacia complex; other species, including Stenotrophomonas maltophilia, Alcaligenes (Achromobacter) xylosoxidans, B. gladioli, and R. pickettii have been recovered from sputum cultures of CF patients as well (2,3). Recently, we showed that a number of unusual bacterial species (including several novel species within the α-Proteobacteria) are also occasionally isolated from CF patients (4). Infection with mucoid P. aeruginosa and members of the B. cepacia complex is associated with increased illness and death in CF patients (5–7), but the clinical importance of infection with these other species is less clear.
The genus Ralstonia was proposed in 1995 (8). Since its creation, the taxonomy of the genus has expanded to include 11 species, which are R. pickettii, R. solanacearum, R. eutropha, R. gilardii, R. paucula, R. basilensis, R. oxalatica, R. mannitolilytica, R. taiwanensis, R. campinensis, and R. metallidurans (8–14). Ralstonia spp. are isolated from a wide variety of ecologic niches, including plants and soils contaminated with heavy metals. R. pickettii has been associated with nosocomial outbreaks caused by contaminated solutions used for patient care and with pseudoepidemics caused by contaminated solutions in the diagnostic laboratory (15–21). Several hospital-associated outbreaks attributed to R. mannitolilytica (formerly known as R. pickettii biovar 3 or P. thomasii) have been described (12,22,23). R. paucula and R. gilardii have only sporadically been isolated from human clinical samples, including cerebrospinal fluid, bone marrow, wounds, and the respiratory tract (9,10). A complete assessment of the frequency of human infection due to Ralstonia species is confounded by the difficulty in accurate species identification by using standard microbiologic techniques. Indeed, these species are frequently misidentified as P. fluorescens or B. cepacia complex (12,24–26).
We describe the occurrence of several Ralstonia species in the respiratory secretions of CF patients. We also describe the development and evaluation of two polymerase chain reaction (PCR) assays for rapid, accurate identification of R. pickettii and R. mannitolilytica.
Bacterial Strains and Study Population
Since early 1997, the Burkholderia cepacia Research Laboratory and Repository (University of Michigan, Ann Arbor, MI) has received more than 4,000 bacterial isolates, collected from CF patients receiving care in 145 CF treatment centers in 130 U. S. cities. Isolates received were tentatively identified by the referring microbiology laboratory as B. cepacia complex or a related species or were not identified to the species level. From these isolates, we identified 42 Ralstonia isolates obtained from 38 patients who had received care in 19 treatment centers in 18 U. S. cities. The type and reference strains of Ralstonia, Pandoraea, Burkholderia, Alcaligenes, and Bordetella species have been described (9–14). These strains were obtained from the BCCM/LMG-Bacteria Collection (Laboratorium voor Microbiologie, Universiteit Gent, Belgium) or were provided by D. Henry (University of British Columbia, Vancouver, Canada). All isolates were grown aerobically on Mueller-Hinton broth (Becton, Dickinson and Company, Cockeysville, MD) supplemented with 1.8% (wt/vol) agar and incubated at 32°C.
Species Identification
We used a polyphasic approach to identify all isolates, including biochemical tests, 16S ribosomal (r)DNA-based PCR assays and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of whole-cell proteins. Biochemical tests (determination of oxidase, lysine decarboxylase, and o-nitrophenyl-β-D-galactoside activity; growth on B. cepacia selective agar; and oxidation-fermentation of sucrose) were performed as described (27). SDS-PAGE of whole-cell proteins was performed as described (9,10), and isolates were identified by comparison with a database containing protein profiles of all Ralstonia species. We used 16S rDNA-based PCR assays (28) to determine whether or not isolates belonged to the genera Burkholderia or Ralstonia or to the B. cepacia complex.
Genotyping of Serial Isolates
Multiple isolates from a single patient were genotyped by randomly amplified polymorphic DNA (RAPD) genotyping as described (29). We digitized gel images with a GelDoc2000 gel analyzer (Bio-Rad Laboratories, Hercules, CA) and stored them as tagged image files. After normalization with the molecular weight marker, patterns were analyzed with Molecular Analyst Fingerprinting Plus software (Bio-Rad Laboratories). Similarities between patterns were calculated by using Pearson’s product-moment correlation coefficient. We considered isolates to belong to the same genotype if they shared 90% or more similarity.
Development of Primers for Species-Specific PCR Assays
We retrieved 16S rDNA sequences of all Ralstonia spp. and representatives of related genera from the GenBank database, using the MegAlign software package (DNASTAR Inc., Madison, WI) to align the sequences. Based on this alignment, we developed primers specific for R. pickettii and R. mannitolilytica: Rp-F1 (5´-ATGATCTAGCTTGCTAGATTGAT-3´) and Rp-R1 (5´-ACTGATCGTCGCCTTGGTG-3´) (forward and reverse primers for the identification of R. pickettii) and Rm-F1 (5´-GGGAAAGCTTGCTTTCCTGCC-3´) and Rm-R1 (5´-TCCGGGTATTAACCAGAGCCAT-3´) (forward and reverse primers for the identification of R. mannitolilytica).
Polymerase Chain Reaction
DNA was prepared as described (30). PCR assays were performed in 25-µL reaction mixtures, containing 2 µL DNA solution, 1U Taq polymerase (GIBCO Invitrogen Corp., Gaithersburg, MD), 250 mM (each) deoxynucleotide triphosphate (GIBCO Invitrogen Corp.), 1.5 mM MgCl2, 1x PCR buffer (GIBCO Invitrogen Corp.), and 20 pmol of each oligonucleotide primer. Amplification was carried out with a PTC-100 programmable thermal cycler (Labtrade Inc., Miami, FL). After initial denaturation for 2 min at 94°C, 30 amplification cycles were completed, each consisting of 1 min at 94°C, 1 min at 55°C (for identifying R. pickettii) or 57°C (for identifying R. mannitolilytica), and 1 min 30 s at 72°C. A final extension of 10 min at 72°C was applied. Negative control PCRs with all reaction mixture components except template DNA were used for every experiment.
Evaluation of the PCR Assays
For evaluating PCR assays, we tested 152 isolates, including 79 Ralstonia isolates (both clinical isolates and reference strains) and 73 isolates representing phylogenetically related species that may be found in sputum cultures of CF patients. Isolates tested were, as follows: R. pickettii (27 isolates), R. mannitolilytica (34), R. gilardii (4), R. paucula (2), R. taiwanensis (1), R. basilensis (1), R. eutropha (1), R. oxalatica (1), R. solanacearum (1), R. campinensis (1), R. metallidurans (1), Ralstonia sp. (5), B. cepacia genomovar I (3), B. multivorans (2), B. cepacia genomovar III (7), B. stabilis (2), B. vietnamiensis (2), B. cepacia genomovar VI (5), B. ambifaria (3), B. gladioli (6), B. fungorum (1), Pandoraea apista (5), P. norimbergensis (3), P. pnomenusa (2), P. sputorum (4), P. pulmonicola (2), Alcaligenes xylosoxidans (5), P. aeruginosa (5), S. maltophilia (5), and one isolate each of A. denitrificans, A. piechaudii, A. faecalis, A. ruhlandii, Bordetella avium, B. hinzii, B. trematum, B. bronchiseptica, B. pertussis, B. parapertussis, and B. holmesii.
Species Identification
Isolates were tentatively identified as belonging to the genus Ralstonia if they 1) reacted with primer pair RHG-F/RHG-R (specific for Burkholderia and Ralstonia spp.) (28), 2) showed no lysine decarboxylase and o-nitrophenyl-β-D-galactoside activity, 3) produced no acid from sucrose, and 4) showed oxidase activity. Using these criteria, we identified 42 putative Ralstonia sp. isolates. These isolates were further identified to the species level by using SDS-PAGE of whole-cell proteins. Most isolates (25) were identified as R. mannitolilytica; 9 were identified as R. pickettii. Two isolates were identified as R. gilardii, and another as R. taiwanensis. Five isolates clearly belonged to the genus Ralstonia but could not be classified into one of the known species. Pending further investigations, these isolates were classified as Ralstonia sp.
Genotyping of Serial Isolates
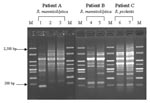
We identified two patients (A and B) who were sputum-culture positive for R. mannitolilytica and one patient (C) who was culture positive for R. pickettii on more than one occasion. The three R. mannitolilytica isolates cultured from patient A were recovered over a period of >2 years. RAPD genotyping indicated that the first isolate clearly differed from the two isolates recovered subsequently; the latter two isolates (recovered 20 months apart) were the same genotype (Figure 1). Similarly, the two R. mannitolilytica isolates recovered from patient B (cultured 8 weeks apart) were the same genotype, as were the two R. pickettii isolates recovered from patient C (cultured 6 weeks apart) (Figure 1).
Primer Design
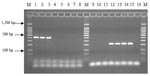
Alignment of 16S rRNA gene sequences of Ralstonia sp. available in GenBank showed similarity values >93.1% and >98.2% within the species R. pickettii and R. mannitolilytica, respectively. Identity of sequences between these two species ranged from 89.9%–96.8%. Several species-level sequence signatures were detected and were incorporated into the species-specific primers Rp-F1 and Rp-R1 (forward and reverse primer for R. pickettii) and Rm-F1 and Rm-R1 (forward and reverse primer for R. mannitolilytica). PCR with these primers resulted in the amplification of fragments of 210 bp and 398 bp, respectively (Figure 2). Each of the 152 strains included in this study was examined by PCR with the primer pairs described (Table).
The occurrence and clinical role of Ralstonia sp. in the respiratory secretions of persons with CF have not been systematically investigated because of the rapidly changing taxonomy of the genus Ralstonia and the absence of rapid, reliable methods for species identification. We used a polyphasic approach to identify Ralstonia sp. in sputum cultures of CF patients and developed two PCR assays for identifying R. pickettii and R. mannitolilytica.
Previous reports describing the bacterial flora of the respiratory tract of CF patients have focused mainly on P. aeruginosa and B. cepacia complex organisms (3,5,31); reports describing the presence of Ralstonia species in sputum cultures of CF patients are scarce and often anecdotal. In a prospective study, Burns et al. (2) isolated R. pickettii from only 2 of 559 patients. More recently, we have shown that other Ralstonia species, including R. mannitolilytica, R. taiwanensis, and R. gilardii, can also be isolated from the respiratory secretions of CF patients (4). In this study, we identified Ralstonia species recovered from sputum cultures of 38 CF patients. Collectively, these data indicate that the prevalence of Ralstonia sp. in the CF population is rather low. However, because we did not specifically survey all referring laboratories for all Ralstonia species that may have been recovered from CF specimens, we were not able to define a more precise prevalence of Ralstonia sp. in the CF population.
Our data do not provide evidence for patient-to-patient spread of Ralstonia sp. because no clustering of cases occurred within centers or geographic regions (data not shown). However, we were able to document persistent colonization with Ralstonia species in three patients. Patient A’s infection is particularly interesting. In this patient, an initial R. mannitolilytica strain was apparently replaced with another strain, which then persisted for >20 months. However, the bacterial and host factors involved in infection by more that one R. mannitolilytica strain or with chronic colonization remain to be defined.
Five Ralstonia isolates could not be identified to the species level. 16S rDNA PCR and SDS-PAGE of whole-cell proteins clearly indicated that these isolates belong to the genus Ralstonia, suggesting that they may represent novel Ralstonia sp. Further polyphasic taxonomic studies are needed to clarify their status. The finding of R. mannitolilytica, R. gilardii, R. taiwanensis, and possible novel Ralstonia species in respiratory secretions of CF patients suggests that these organisms may be emerging human pathogens and again highlights the fact that the bacterial biodiversity in the respiratory tract of CF patients has thus far been underestimated (4).
Of the 25 R. mannitolilytica strains identified in this study, 9 were initially identified by the referring laboratory as R. pickettii, 8 as B. cepacia complex, 6 as Burkholderia sp., 1 as B. gladioli, and 1 as P. fluorescens. Of the 9 R. pickettii strains identified, 3 were identified by the referring laboratory as R. pickettii, 2 as Burkholderia sp., 1 as Pseudomonas sp., 1 as B. cepacia complex, and 2 isolates as unidentified. The R. gilardii and R. taiwanensis isolates were received as B. cepacia complex and S. maltophilia, respectively. Most (81%) of these isolates were capable of growth on B. cepacia selective agar. These observations reiterate that identification of these species is not straightforward and that their misidentification as other CF pathogens, such as B. cepacia complex, is not uncommon. Such misidentification has an important impact on infection control in CF since the efficiency of these measures depends on accurate identification of the microorganisms involved. Infection-control policies, particularly those recommended to prevent interpatient spread of B. cepacia complex, have a tremendous impact on the quality of life of CF patients (6,7). To enhance accurate identification of CF pathogens, several PCR assays have been developed recently (28,30,32–35). We sought to design similar PCR tests to allow the identification of R. pickettii and R. mannitolilytica based on species-level signature sequences in the 16S rRNA gene. By comparing available R. pickettii and R. mannitolilytica 16S rRNA gene sequences with sequences from other Ralstonia species and representatives of the phylogenetically closely related genera Burkholderia and Pandoraea, we identified several regions that showed sufficient diversity to allow the design of primer pairs Rp-F1/Rp-R1 and Rm-F1/Rm-R1, permitting the sensitive and specific identification of R. pickettii and R. mannitolilytica, respectively (Table).
The results of our study indicate that a number of Ralstonia species can be isolated from sputum cultures of CF patients. The correct identification of these species presents a challenge for diagnostic microbiology laboratories. Our study supports the use of genotypic methods to augment routine phenotypic evaluation. The combined use of the two PCR assays described will allow the identification of most Ralstonia species encountered in sputum cultures of CF patients. Most importantly, the use of these assays will substantially reduce the misidentification of R. pickettii and R. mannitolilytica as B. cepacia complex. These tests will be a valuable adjunct in the evaluation of CF sputum culture isolates and will allow more precise study of the prevalence and natural history of human infection by these emerging pathogens.
Dr. Coenye is a postodoctoral research fellow in the Department of Pediatrics and Communicable Diseases at the University of Michigan. His major research interests are the identification, biodiversity, and molecular epidemiology of bacteria associated with pulmonary infections in cystic fibrosis patients.
Acknowledgments
We thank T. Spilker and A. Martin for excellent technical assistance.
This work was supported by a grant from the Cystic Fibrosis Foundation (United States) (to JJL). TC is supported by the Caroll Haas Research Fund in Cystic Fibrosis.
References
- Burns JL, Emerson J, Stapp JR, Yim DL, Krzewinski J, Louden L, Microbiology of sputum from patients at cystic fibrosis centers in the United States. Clin Infect Dis. 1998;27:158–63. DOIPubMedGoogle Scholar
- Gilligan PH. Microbiology of airway disease in patients with cystic fibrosis. Clin Microbiol Rev. 1991;4:35–51.PubMedGoogle Scholar
- Coenye T, Vandamme P, LiPuma JJ. Characterisation of unusual bacteria isolated from CF sputum [abstract]. Pediatr Pulmonol. 2001;Suppl 22:297.PubMedGoogle Scholar
- Govan JRW, Deretic V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev. 1996;60:539–74.PubMedGoogle Scholar
- LiPuma JJ. Burkholderia cepacia. Management issues and new insights. Clin Chest Med. 1998;19:473–86. DOIPubMedGoogle Scholar
- LiPuma JJ. Burkholderia cepacia epidemiology and pathogenesis: implications for infection control. Curr Opin Pulm Med. 1998;4:337–441. DOIPubMedGoogle Scholar
- Yabuuchi E, Kosako Y, Yano I, Hotta H, Nishiuchi Y. Transfer of two Burkholderia and an Alcaligenes species to Ralstonia gen. nov.: proposal of Ralstonia pickettii (Ralston, Palleroni and Doudoroff 1973) comb. nov., Ralstonia solanacearum (Smith 1896) comb. nov. and Ralstonia eutropha (Davis 1969) comb. nov. Microbiol Immunol. 1995;39:897–904.PubMedGoogle Scholar
- Coenye T, Falsen E, Vancanneyt M, Hoste B, Govan JRW, Kersters K, Classification of some Alcaligenes faecalis-like isolates from the environment and human clinical samples as Ralstonia gilardii sp. nov. Int J Syst Bacteriol. 1999;49:405–13.PubMedGoogle Scholar
- Vandamme P, Goris J, Coenye T, Hoste B, Janssens D, Kersters K, Assignment of Centers for Disease Control group IVc-2 to the genus Ralstonia as Ralstonia paucula sp. nov. Int J Syst Bacteriol. 1999;49:663–9.PubMedGoogle Scholar
- Sahin N, Isik K, Tamer AU, Goodfellow M. Taxonomic position of “Pseudomonas oxalaticus” strain ox14T (DSM 1105T) (Khambata and Bhat, 1953) and its description in the genus Ralstonia as Ralstonia oxalatica comb. nov. Syst Appl Microbiol. 2000;23:206–9.PubMedGoogle Scholar
- De Baere T, Steyaert S, Wauters G, De Vos P, Goris J, Coenye T, Classification of Ralstonia picketii biovar 3/’thomasii’ strains (Pickett 1994) and of new isolates related to nosocomial recurrent meningitis as Ralstonia mannitolytica sp. nov. Int J Syst Evol Microbiol. 2001;51:547–58.PubMedGoogle Scholar
- Chen WM, Laevens S, Lee TM, Coenye T, De Vos P, Mergeay M, Ralstonia taiwanensis sp. nov., isolated from root nodules of Mimosa species and sputum of a cystic fibrosis patient. Int J Syst Evol Microbiol. 2001;51:1729–35.PubMedGoogle Scholar
- Goris J, De Vos P, Coenye T, Hoste B, Janssens D, Brim H, Classification of metal-resistant bacteria from industrial biotopes as Ralstonia campinensis sp. nov., Ralstonia metallidurans sp. nov. and Ralstonia basilensis Steinle et al. 1998 emend. Int J Syst Evol Microbiol. 2001;51:1773–82.PubMedGoogle Scholar
- Riley PS, Weaver RE. Recognition of Pseudomonas pickettii in the clinical laboratory: biochemical characterization of 62 strains. J Clin Microbiol. 1975;1:61–4.PubMedGoogle Scholar
- Labarca JA, Trick WE, Peterson CL, Carson LA, Holt SC, Arduino MJ, A multistate nosocomial outbreak of Ralstonia pickettii colonization associated with an intrinsically contaminated respiratory care solution. Clin Infect Dis. 1999;29:1281–6. DOIPubMedGoogle Scholar
- Lacey S, Want SV. Pseudomonas pickettii infections in a paediatric oncology unit. J Hosp Infect. 1991;17:45–51. DOIPubMedGoogle Scholar
- Fernandez C, Wilhelmi I, Andradas E, Gaspar C, Gomez J, Romero J, Nosocomial outbreak of Burkholderia pickettii infection due to a manufactured intravenous product used in three hospitals. Clin Infect Dis. 1996;22:1092–5.PubMedGoogle Scholar
- McNeil MM, Solomon SL, Anderson RL, Davis BJ, Spengler RF, Reisberg BE, Nosocomial Pseudomonas pickettii colonization associated with a contaminated respiratory therapy solution in a special care nursery. J Clin Microbiol. 1985;22:903–7.PubMedGoogle Scholar
- Verschraegen G, Claeys G, Meeus G, Delanghe M. Pseudomonas pickettii as a cause of pseudobacteremia. J Clin Microbiol. 1985;21:278–9.PubMedGoogle Scholar
- Centers for Disease Control and Prevention. Nosocomial Ralstonia pickettii colonization associated with intrinsically contaminated saline solution—Los Angeles, California, 1998. MMWR Morb Mortal Wkly Rep. 1998;47:285–6.PubMedGoogle Scholar
- Phillips I, Eykyn S, Laker M. Outbreak of hospital infection caused by contaminated autoclaved fluids. Lancet. 1972;1:1258–60. DOIPubMedGoogle Scholar
- Costas M, Holmes B, Sloss LL, Heard S. Investigation of a pseudo-outbreak of ‘Pseudomonas thomasii’ in a special-care baby unit by numerical analysis of SDS-PAGE protein patterns. Epidemiol Infect. 1990;105:127–37. DOIPubMedGoogle Scholar
- Henry DA, Mahenthiralingam E, Vandamme P, Coenye T, Speert DP. Phenotypic methods for determining genomovar status of the Burkholderia cepacia complex. J Clin Microbiol. 2001;39:1073–8. DOIPubMedGoogle Scholar
- Kiska DL, Kerr A, Jones MC, Caracciolo JA, Eskridge B, Jordan M, Accuracy of four commercial systems for identification of Burkholderia cepacia and other gram-negative nonfermenting bacilli recovered from patients with cystic fibrosis. J Clin Microbiol. 1996;34:886–91.PubMedGoogle Scholar
- Gilligan P. Pseudomonas and Burkholderia. In: Murray PR, Baron EJ, Pfaller MA, Tenover FC, Yolken RH, editors. Manual of clinical microbiology. 6th edition. Washington: ASM Press; 1995. p. 509–32.
- McMenamin JD, Zaccone TM, Coenye T, Vandamme P, LiPuma JJ. Misidentification of Burkholderia cepacia in US cystic fibrosis treatment centers: an analysis of 1051 recent sputum isolates. Chest. 2000;117:1661–5. DOIPubMedGoogle Scholar
- LiPuma JJ, Dulaney BJ, McMenamin JD, Whitby PW, Stull TL, Coenye T, Development of rRNA-based PCR assays for identification of Burkholderia cepacia complex isolates recovered from cystic fibrosis patients. J Clin Microbiol. 1999;37:3167–70.PubMedGoogle Scholar
- Chen JS, Witzmann KA, Spilker T, Fink RJ, LiPuma JJ. Endemicity and inter-city spread of Burkholderia cepacia genomovar III in cystic fibrosis. J Pediatr. 2001;139:643–9. DOIPubMedGoogle Scholar
- Coenye T, Liu L, Vandamme P, LiPuma JJ. Identification of Pandoraea species by 16S ribosomal DNA-based PCR assays. J Clin Microbiol. 2001;39:4452–5. DOIPubMedGoogle Scholar
- Klinger JD, Thomassen MJ. Occurrence and antimicrobial susceptibility of gram-negative nonfermentative bacilli in cystic fibrosis patients. Diagn Microbiol Infect Dis. 1985;3:149–58. DOIPubMedGoogle Scholar
- Coenye T, Vandamme P, Govan JRW, LiPuma JJ. Taxonomy and identification of the Burkholderia cepacia complex. J Clin Microbiol. 2001;39:3427–36. DOIPubMedGoogle Scholar
- Whitby PW, Pope LC, Carter KB, LiPuma JJ, Stull TL. Species-specific PCR as a tool for the identification of Burkholderia gladioli. J Clin Microbiol. 2000;38:282–5.PubMedGoogle Scholar
- Whitby PW, Carter KB, Burns JL, Royall JA, LiPuma JJ, Stull TL. Identification and detection of Stenotrophomonas maltophilia by rRNA-directed PCR. J Clin Microbiol. 2000;38:4305–9.PubMedGoogle Scholar
- Liu L, Coenye T, Burns JL, Whitby PW, Stull TL, LiPuma JJ. Ribosomal DNA-directed PCR for identification of Achromobacter (Alcaligenes) xylosoxidans recovered from sputum samples from cystic fibrosis patients. J Clin Microbiol. 2002;40:1210–3. DOIPubMedGoogle Scholar
Figures
Table
Cite This ArticleTable of Contents – Volume 8, Number 7—July 2002
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Tom Coenye, Department of Pediatrics and Communicable Diseases, 8301 MSRB III, Box 0646, 1150 W. Medical Center Dr., Ann Arbor, Michigan 48109-0646, USA; fax: 734-615-4770;
Top