Volume 14, Number 2—February 2008
Research
Unexpected Occurrence of Plasmid-Mediated Quinolone Resistance Determinants in Environmental Aeromonas spp.
Figure 2
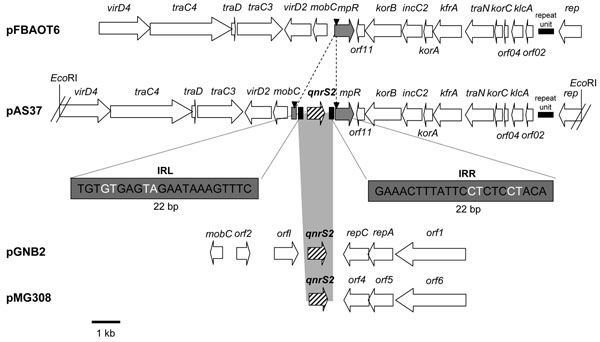
Figure 2. Genetic environments of the qnrS2 gene in plasmid p37 from Aeromonas punctata 37 and comparison with related plasmid structures. Plasmid pFBAOT6 is from A. punctata from the United Kingdom (23); plasmids pGNB2 and pMG308 are from a wastewater treatment plant from Germany (unknown bacterial reservoir) (24) and from a non-Typhi Salmonella clinical isolate from the United States (25), respectively. Recombinant plasmid pAS37 has been obtained from our study. Open reading frames (ORFs) are indicated by horizontal arrows. The right and left inverted repeats (IRR and IRL) are indicated, and duplication sites (CCTCC) are represented by black triangles. The EcoRI- restriction sites that have been used for cloning experiments are indicated. The identified mobile insertion cassette element is bracketed by IRL and IRR of 22-bp size (bases in black are identical, and bases in white are different).
References
- Hooper DC. Emerging mechanisms of fluoroquinolone resistance. Emerg Infect Dis. 2001;7:337–41.PubMedGoogle Scholar
- Ruiz J. Mechanisms of resistance to quinolones: target alterations, decreased accumulation and DNA gyrase protection. J Antimicrob Chemother. 2003;51:1109–17. DOIPubMedGoogle Scholar
- Martinez-Martinez L, Pascual A, Jacoby GA. Quinolone resistance from a transferable plasmid. Lancet. 1998;351:797–9. DOIPubMedGoogle Scholar
- Tran JH, Jacoby GA, Hooper DC. Interaction of the plasmid-encoded quinolone resistance protein Qnr with Escherichia coli DNA gyrase. Antimicrob Agents Chemother. 2005;49:118–25. DOIPubMedGoogle Scholar
- Tran JH, Jacoby GA, Hooper DC. Interaction of the plasmid-encoded quinolone resistance protein QnrA with Escherichia coli topoisomerase IV. Antimicrob Agents Chemother. 2005;49:3050–2. DOIPubMedGoogle Scholar
- Nordmann P, Poirel L. Emergence of plasmid-mediated resistance to quinolones in Enterobacteriaceae. J Antimicrob Chemother. 2005;56:463–9. DOIPubMedGoogle Scholar
- Martinez-Martinez L, Pascual A, Garcia I, Tran J, Jacoby GA. Interaction of plasmid and host quinolone resistance. J Antimicrob Chemother. 2003;51:1037–9. DOIPubMedGoogle Scholar
- Robicsek A, Jacoby GA, Hooper DC. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect Dis. 2006;6:629–40. DOIPubMedGoogle Scholar
- Hata M, Suzuki M, Matsumoto M, Takahashi M, Sato K, Ibe S, Cloning of a novel gene for quinolone resistance from a transferable plasmid in Shigella flexneri 2b. Antimicrob Agents Chemother. 2005;49:801–3. DOIPubMedGoogle Scholar
- Jacoby GA, Walsh KE, Mills DM, Walker VJ, Oh H, Robicsek A, qnrB, another plasmid-mediated gene for quinolone resistance. Antimicrob Agents Chemother. 2006;50:1178–82. DOIPubMedGoogle Scholar
- Poirel L, Rodriguez-Martinez JM, Mammeri H, Liard A, Nordmann P. Origin of plasmid-mediated quinolone resistance determinant QnrA. Antimicrob Agents Chemother. 2005;49:3523–5. DOIPubMedGoogle Scholar
- Cattoir V, Poirel L, Mazel D, Soussy CJ, Nordmann P. Vibrio splendidus as the source of plasmid-mediated QnrS-like quinolone resistance determinants. Antimicrob Agents Chemother. 2007;51:2650–1. DOIPubMedGoogle Scholar
- Poirel L, Liard A, Rodriguez-Martinez JM, Nordmann P. Vibrionaceae as a possible source of Qnr-like quinolone resistance determinants. J Antimicrob Chemother. 2005;56:1118–21. DOIPubMedGoogle Scholar
- Dortet L, Legrand P, Soussy CJ, Cattoir V. Bacterial identification, clinical significance, and antimicrobial susceptibilities of Acinetobacter ursingii and Acinetobacter schindleri, two frequently misidentified opportunistic pathogens. J Clin Microbiol. 2006;44:4471–8. DOIPubMedGoogle Scholar
- Yanez MA, Catalan V, Apraiz D, Figueras MJ, Martinez-Murcia AJ. Phylogenetic analysis of members of the genus Aeromonas based on gyrB gene sequences. Int J Syst Evol Microbiol. 2003;53:875–83. DOIPubMedGoogle Scholar
- Cattoir V, Poirel L, Rotimi V, Soussy CJ, Nordmann P. Multiplex PCR for detection of plasmid-mediated quinolone resistance qnr genes in ESBL-producing enterobacterial isolates. J Antimicrob Chemother. 2007;60:394–7. DOIPubMedGoogle Scholar
- Poirel L, Leviandier C, Nordmann P. Prevalence and genetic analysis of plasmid-mediated quinolone resistance determinants QnrA and QnrS in Enterobacteriaceae isolates from a French university hospital. Antimicrob Agents Chemother. 2006;50:3992–7. DOIPubMedGoogle Scholar
- Goni-Urriza M, Arpin C, Capdepuy M, Dubois V, Caumette P, Quentin C. Type II topoisomerase quinolone resistance-determining regions of Aeromonas caviae, A. hydrophila, and A. sobria complexes and mutations associated with quinolone resistance. Antimicrob Agents Chemother. 2002;46:350–9. DOIPubMedGoogle Scholar
- Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; 17th informational supplement M100–S17. Wayne (PA): The Institute; 2007.
- Aubron C, Poirel L, Ash RJ, Nordmann P. Carbapenemase-producing Enterobacteriaceae, US rivers. Emerg Infect Dis. 2005;11:260–4.PubMedGoogle Scholar
- Casas C, Anderson EC, Ojo KK, Keith I, Whelan D, Rainnie D, Characterization of pRAS1-like plasmids from atypical North American psychrophilic Aeromonas salmonicida. FEMS Microbiol Lett. 2005;242:59–63. DOIPubMedGoogle Scholar
- Kieser T. Factors affecting the isolation of CCC DNA from Streptomyces lividans and Escherichia coli. Plasmid. 1984;12:19–36. DOIPubMedGoogle Scholar
- Rhodes G, Parkhill J, Bird C, Ambrose K, Jones MC, Huys G, Complete nucleotide sequence of the conjugative tetracycline resistance plasmid pFBAOT6, a member of a group of IncU plasmids with global ubiquity. Appl Environ Microbiol. 2004;70:7497–510. DOIPubMedGoogle Scholar
- Bonemann G, Stiens M, Puhler A, Schlueter A. Mobilizable IncQ-related plasmid carrying a new quinolone resistance gene, qnrS2, isolated from the bacterial community of a wastewater treatment plant. Antimicrob Agents Chemother. 2006;50:3075–80. DOIPubMedGoogle Scholar
- Gay K, Robicsek A, Strahilevitz J, Park CH, Jacoby GA, Barrett TJ, Plasmid-mediated quinolone resistance in non-Typhi serotypes of Salmonella enterica. Clin Infect Dis. 2006;43:297–304. DOIPubMedGoogle Scholar
- De Palmenaer D, Vermeiren C, Mahillon J. IS231-MIC231 elements from Bacillus cereus sensu lato are modular. Mol Microbiol. 2004;53:457–67. DOIPubMedGoogle Scholar
- Chatzipanagiotou S, Ioannidou V, Ioannidis A, Nicolaou C, Papavasileiou E, Chaniotaki S, Absence of the plasmid-mediated quinolone resistance qnrA gene among Campylobacter jejuni clinical isolates from Greece. Int J Antimicrob Agents. 2005;26:261–2. DOIPubMedGoogle Scholar
- Kehrenberg C, Friederichs S, de Jong A, Michael GB, Schwarz S. Identification of the plasmid-borne quinolone resistance gene qnrS in Salmonella enterica serovar Infantis. J Antimicrob Chemother. 2006;58:18–22. DOIPubMedGoogle Scholar
- Hopkins KL, Wootton L, Day MR, Threlfall EJ. Plasmid-mediated quinolone resistance determinant qnrS1 found in Salmonella enterica strains isolated in the UK. J Antimicrob Chemother. 2007;59:1071–5. DOIPubMedGoogle Scholar
- Poirel L, Nguyen TV, Weintraub A, Leviandier C, Nordmann P. Plasmid-mediated quinolone resistance determinant qnrS in Enterobacter cloacae. Clin Microbiol Infect. 2006;12:1021–3. DOIPubMedGoogle Scholar
- Chen YT, Shu HY, Li LH, Liao TL, Wu KM, Shiau YR, Complete nucleotide sequence of pK245, a 98-kilobase plasmid conferring quinolone resistance and extended-spectrum-beta-lactamase activity in a clinical Klebsiella pneumoniae isolate. Antimicrob Agents Chemother. 2006;50:3861–6. DOIPubMedGoogle Scholar
- Wu JJ, Ko WC, Tsai SH, Yan JJ. Prevalence of plasmid-mediated quinolone resistance determinants QnrA, QnrB, and QnrS among clinical isolates of Enterobacter cloacae in a Taiwanese hospital. Antimicrob Agents Chemother. 2007;51:1223–7. DOIPubMedGoogle Scholar
- Cavaco LM, Hansen DS, Friis-Moller A, Aarestrup FM, Hasman H, Frimodt-Moller N. First detection of plasmid-mediated quinolone resistance (qnrA and qnrS) in Escherichia coli strains isolated from humans in Scandinavia. J Antimicrob Chemother. 2007;59:804–5. DOIPubMedGoogle Scholar
- Rhodes G, Huys G, Swings J, McGann P, Hiney M, Smith P, Distribution of oxytetracycline resistance plasmids between aeromonads in hospital and aquaculture environments: implication of Tn1721 in dissemination of the tetracycline resistance determinant tetA. Appl Environ Microbiol. 2000;66:3883–90. DOIPubMedGoogle Scholar
- Schmidt AS, Bruun MS, Dalsgaard I, Larsen JL. Incidence, distribution, and spread of tetracycline resistance determinants and integron-associated antibiotic resistance genes among motile aeromonads from a fish farming environment. Appl Environ Microbiol. 2001;67:5675–82. DOIPubMedGoogle Scholar
- Young HK. Antimicrobial resistance spread in aquatic environments. J Antimicrob Chemother. 1993;31:627–35. DOIPubMedGoogle Scholar
- Poirel L, Cattoir V, Soares A, Soussy CJ, Nordmann P. Novel Ambler class A β-lactamase LAP-1 and its association with the plasmid-mediated quinolone resistance determinant QnrS1. Antimicrob Agents Chemother. 2006;51:631–7. DOIPubMedGoogle Scholar
- Kümmerer K. Resistance in the environment. J Antimicrob Chemother. 2004;54:311–20. DOIPubMedGoogle Scholar
- Beaber JW, Hochhut B, Waldor MK. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature. 2004;427:72–4. DOIPubMedGoogle Scholar