Volume 14, Number 2—February 2008
Research
Unexpected Occurrence of Plasmid-Mediated Quinolone Resistance Determinants in Environmental Aeromonas spp.
Cite This Article
Citation for Media
Abstract
We searched for plasmid-mediated quinolone resistance determinants of the Qnr type in several water samples collected at diverse locations from the Seine River (Paris, France). The qnrS2 genes were identified from Aeromonas punctata subsp. punctata and A. media. The qnrS2 gene was located on IncU-type plasmids in both isolates, which resulted in increased MIC values of quinolones and fluoroquinolones, once they were transferred into Escherichia coli. The qnrS2 gene identified in A. punctata was part of novel genetic structure corresponding to a mobile insertion cassette element. This identification of plasmid-mediated qnr genes outside Enterobacteriaceae underlines a possible diffusion of those resistance determinants within gram-negative rods.
Quinolones are broad-spectrum antibacterial agents used in human and veterinary medicine. Their extensive use has been associated with a rising level of quinolone resistance (1). The 2 main mechanisms of quinolone resistance are chromosomally encoded, either a modification of the quinolone targets with changes of DNA gyrase (gyrA) and/or of topoisomerase IV (parC) genes; or a decreased intracellular concentration due to impermeability of the membrane or to overexpression of efflux pump systems (2). Plasmid-mediated quinolone resistance was first identified in a Klebsiella pneumoniae clinical isolate from the United States (3). It is mediated by a 218-aa protein, Qnr (lately termed QnrA), which belongs to the pentapeptide repeat family of proteins that protects DNA from quinolone binding to topoisomerases (4,5). QnrA confers resistance to quinolones such as nalidixic acid and increases MICs of fluoroquinolones up to 32-fold in Escherichia coli (6). In addition, it enhances selection of associated chromosome-encoded quinolone resistance determinants that confer additional resistance to fluoroquinolones (7). The QnrA determinants have been reported worldwide in many enterobacterial species, and 6 of them are known so far (QnrA1 to QnrA6) (8). Other plasmid-mediated quinolone resistance determinants, QnrB (QnrB1 to QnrB10) and QnrS (QnrS1 and QnrS2), have been identified in enterobacterial species, sharing 41% and 60% amino acid identity with QnrA, respectively (8–10). The plasmid-mediated qnr genes have been identified so far only in Enterobacteriaceae (6,8). Recent findings indicated that those genes originate from environmental gram-negative bacterial species, such as Shewanella algae, the progenitor of the qnrA genes (11), and Vibrio splendidus, the progenitor of qnrS genes (12). We have shown that many Vibrionaceae species may harbor chromosome-encoded qnr-type genes (13).
To further evaluate the spread of plasmid-mediated resistance determinants in the environment, we have searched for those genes in water samples drawn from the Seine River in Paris, France. We identified QnrS determinants in Aeromonas species in uncommon genetic environments.
Water Sampling and Screening for qnr Genes
Water samples (40 mL each) were collected in November 2006, ≈0.2 m below the water surface, by immersion of 50-mL sterile, screw-capped tubes. Six distinct urban sites located on the Seine River in Paris were sampled: center (site 1), north (site 2), east (site 3), south (site 4), and west (sites 5 and 6). The Seine crosses the city of Paris (2. 4 million inhabitants) from southeast to northwest.
Samples were stored at 4°C before DNA extraction. To estimate the total number of viable bacteria, we diluted all samples in 0.85% saline solution and plated them (0.2 mL) on chromogenic URISelect4 agar (Bio-Rad, Marnes-la-Coquette, France) without any antimicrobial agent, which allowed visual differentiation of gram-negative species. After enrichment of 1 mL of water sample in 10 mL of trypticase soy (TS) broth for 48 h at 30°C and 37°C, qnrA-, qnrB- and qnrS genes were detected by using a multiplex PCR-based strategy (see below).
Isolation and Identification of qnr-Positive Isolates
To identify the qnr-positive isolates, 1 mL of each qnr-positive water sample was grown in 10 mL of TS broth containing nalidixic acid (16 mg/L) for 24–48 h at 30°C and 37°C. After dilution in 0.85% NaCl, each broth was plated (0.2 mL) on chromogenic URISelect4 agar and 5% horse-blood TS agar, both containing nalidixic acid (16 mg/L). Finally, each colony was tested by multiplex PCR for detection of qnr genes (see below). Further identification of qnr-positive isolates was performed by conventional biochemical techniques (API-20E and API-NE systems [bioMérieux, Marcy-l’Etoile, France]), and by sequencing of 16S rDNA and gyrB genes, as previously described (14,15).
Isolation of Total DNA and PCR Amplification
Whole-cell DNA was extracted from water samples by using QIAamp DNA Mini Kit (QIAGEN, Courtaboeuf, France), according to the manufacturer’s recommendations. Whole-cell DNA from isolated colonies identified after culture was extracted by using the boiling technique, which includes a heating step at 100°C of a single colony in a volume of 100 μL of distilled water followed by centrifugation of the cell suspension.
Screening of the qnrA-, qnrB-, and qnrS genes in water samples and from isolated colonies was conducted by using a multiplex PCR-based technique able to amplify the known Qnr variants, as previously described (16). E. coli Lo, K. pneumoniae B1, and E. coli S7 strains were used as qnrA-, qnrB-, and qnrS-positive controls, respectively (16,17). PCR amplicons were sequenced on both strands (see below). The chromosome-encoded quinolone-resistance determining regions (QRDRs) of gyrA and parC genes were sequenced after PCR amplification by using whole-cell DNA of qnr-positive isolates, as previously described (18).
PCR assays were also performed by using standard conditions of amplification (17) to detect genes coding for the replication protein of IncU-type plasmids (rep gene) and the tetracycline resistance determinant (encoded by the tetA gene). The primers, designed for this study, are as follows: for rep, Rep-IncU-F (5′-CTGGCTGAAATGCTGTTGCC-3′) and Rep-IncU-R (5′-GCTTCATAGGCTTCACGCTC-3′) to give a 1,199-bp product, and for tetA, TetA-1 (5′-GTGAAACCCAACAGACCCC-3′) and TetA-2 (5′-TCAGCGATCGGCTCGTTGC-3′) to give a 589-bp product.
Antimicrobial Drug–Susceptibility Testing
The antimicrobial drug susceptibility of Aeromonas isolates was first determined by the disk diffusion technique on Mueller-Hinton (MH) agar plates according to Clinical and Laboratory Standards Institute guidelines (19). The disks were supplied by Bio-Rad Laboratories, and the following antimicrobial agents were tested: amoxicillin (25 μg), ticarcillin (75 μg), amoxicillin-clavulanate (20/10 μg), ticarcillin-clavulanate (75/10 μg), piperacillin (75 µg), piperacillin-tazobactam (75/10 μg), cephalothin (30 μg), cefuroxime (30 μg), cefoxitin (30 μg), cefotaxime (30 μg), ceftazidime (30 μg), aztrem (30 μg), latamoxef (30 μg), cefepime (30 μg), imipenem (10 μg), kanamycin (30 IU), tobramycin (10 μg), gentamicin (15 μg), netilmicin (30 μg), amikacin (30 μg), nalidixic acid (10 μg), norfloxacin (5 μg), ofloxacin (5 μg), ciprofloxacin (5 μg), chloramphenicol (30 μg), tetracycline 30 (IU), fosfomycin (50 μg), rifampicin (30 μg), sulfonamide (200 μg), trimethoprim (5 μg), and colistin (50 μg).
Thus, MICs of quinolones and fluoroquinolones were determined by using the E test technique, according to the manufacturer’s recommendations (AB Biodisk, Solna, Sweden). MIC breakpoints were retained for determining susceptibility and resistance ranges to nalidixic acid and ciprofloxacin were <8 and >32 mg/L and <1 and >4 mg/L, respectively, as recommended for Enterobacteriaceae (19).
Transfer of Resistance and Plasmid Analysis
Transfer of the plasmid-mediated quinolone resistance markers from qnr-positive Aeromonas isolates to E. coli TOP10 was attempted by using electroporation and conjugation techniques (20). E. coli TOP10 and azide-resistant E. coli J53 strains were recipient strains for transformation and conjugation experiments, respectively (20). Conjugation experiments were performed at 22°C and 37°C, as previously described (21). Transformants and transconjugants were selected on MH agar plates containing nalidixic acid (3 mg/L) only or containing azide (100 mg/L) plus nalidixic acid (6 mg/L), respectively. Plasmid extraction was performed from each qnr-positive isolate and its corresponding transformants by using the Kieser technique (22). Plasmid sizes were determined by electrophoresis on an agarose gel and comparison with sizes of reference plasmids (164, 66, 38, and 7 kb) of E. coli NCTC 50192, as previously described (20). All transformants were confirmed to be qnr positive by mulitplex PCR (see above).
Cloning experiments were performed with EcoRI-restricted whole-cell DNA of Aeromonas isolates 37 and 42, followed by ligation of DNA fragments in the EcoRI-site of cloning vector pBK-CMV. Recombinant plasmids were then transformed by electroporation into E. coli TOP10 electrocompetent cells. E. coli TOP10 harboring recombinant plasmids were selected on MH agar plates containing kanamycin (30 mg/L) and nalidixic acid (3 mg/L). All clones were tested as carrying qnr gene by multiplex PCR (see above), and cloned fragments of recombinant plasmids were sequenced by primer walking (see below).
Sequencing and Bioinformatic Analysis
PCR products, purified with a Qiaquick PCR Purification Kit (QIAGEN), and recombinant plasmids were sequenced by using an Applied Biosystems sequencer (ABI377, Foster City, CA, USA). The nucleotide sequences and the deduced protein sequences were analyzed with BLAST software (www.ncbi.nlm.nih.gov/BLAST).
QnrS2 Determinant from Aeromonas spp.
Samples extracted from 2 of the 6 sites (sites 3 and 4) from the Seine River in autumn 2006 were PCR positive for qnrS genes. Sequencing of the amplicons identified a qnrS2 gene in both cases. Further samples were therefore collected 1 week later from the same collecting sites and assessed the permanent occurrence of QnrS2- positive isolates in that river at that time. No qnrA and qnrB genes were detected.
The total number of gram-negative bacteria from water collection varied from 103 to 105 CFU per mL, depending on the sampling site, and included ≈10%–50% of isolates belonging to Aeromonas species (data not shown). The QnrS2-positive isolates were identified as Aeromonas species according to their phenotypic characterization. Strain 37 (site 4, pink colonies on URI4Select agar) and strain 42 (site 3, blue colonies on URI4Select agar) were selected as qnrS2-positive Aeromonas isolates to be further characterized. Genotypic identification was first attempted by sequencing of a ≈1,200-bp fragment of the 16S rDNA gene, but sequence analysis did not allow a precise identification. Isolate A37 was identified as A. trota or A. punctata (formerly A. caviae) (only 1 nt difference) and isolate A42 as A. hydrophila or A. media (only 1 nt change). Because gyrB sequence divergence is greater than that of 16S rDNA, phylogenetic analysis based on gyrB sequence (allowing more reliable identification of members of the genus Aeromonas [15]) identified isolate A37 as A. punctata subsp. punctata and isolate A39 as A. media.
The Aeromonas isolates both displayed a wild-type resistance phenotype to β-lactams with resistance to narrow-spectrum penicillins and remained susceptible to several β-lactam molecules, including broad-spectrum cephalosporins and carbapenems. A. media 42 was also resistant to several aminoglycosides (kanamycin, tobramycin), chloramphenicol, and tetracycline, whereas A. punctata 37 was fully susceptible.
Aeromonas isolates 37 and 42 were resistant to nalidixic acid (MIC >256 mg/L) and to fluoroquinolones (Table). However, A. media 42 exhibited higher resistance levels to fluoroquinolones, with MICs 2- to 8-fold higher than those for A. punctata 37 (Table). Sequence analysis of the QRDR regions of gyrA and parC genes showed that A. punctata 37 had 1 aa substitution, Ser83Ile in GyrA, whereas A. media 42 had 2 aa substitutions, Ser83Ile in GyrA and Ser80Ile in ParC, as compared with the wild-type proteins of Aeromonas species (18).
qnrS2 Gene on IncU-Plasmid Backbone
The plasmid-mediated quinolone resistance QnrS2 determinant was transferred from Aeromonas isolates 37 and 42 to E. coli TOP10 recipient strain by electrotransformation, but repeated conjugation experiments failed. Plasmid analysis identified a single 55-kb plasmid (p37) and a single 20-kb plasmid (p42) from E. coli transformants from A. punctata 37 and A. media 42 isolates, respectively (Figure 1). After they were transferred into E. coli TOP10, plasmids p37 and p42 conferred increased MICs of quinolones and fluoroquinolones (Table). No other antimicrobial resistance marker was carried by those natural plasmids.
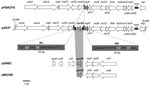
Cloning of EcoRI-restricted DNA from whole-cell DNA of A. punctata 37 produced a recombinant plasmid, pAS37, containing a 19,050-bp insert that contained the qnrS2 gene. Sequencing showed that the qnrS2 gene was located in a plasmid-encoded genetic structure previously identified in an IncU-related plasmid, pFBAOT6 (84,748 bp), isolated from an A. punctata strain recovered from hospital sewage in Kendal (United Kingdom) in 1997 (23). This region consisted of 15 open reading frames (Figure 2). Detailed analysis of pAS37 showed that a fragment of 1,375 bp containing the qnrS2 gene (657 bp) was inserted within the mpR gene coding for a putative zinc metalloprotease (MpR) (Figure 2). Sequencing of the full insert in plasmid pAS37 identified the same rep region as reported from the IncU-related pFBAOT6 plasmid, which indicated that p37 was a member of the IncU incompatibility group. PCR amplification that used specific primers of this rep gene also gave positive results from whole-cell DNA of E. coli (p42) transformant, indicating that p42 also belonged to IncU-type plasmid family. In addition, cloning of EcoRI-restricted DNA from whole-cell DNA of A. media 42 showed that the qnrS2 gene was inserted into the mpR gene in plasmid p42. No tetA gene encoding resistance to tetracycline was detected by PCR in plasmids p37 and p42, whereas it was identified in plasmid pFBAOT6 (23).
qnrS2 Gene as Part of a Mobile Insertion Cassette
The qnrS2 gene was identified inside a 1,375-bp structure bracketed by two 22-bp imperfect inverted repeats (4 mismatches) (Figure 2). This structure ressembled a transposon; however, no transposase encoding gene was associated with the qnrS2 gene, the single gene identified in this genetic structure. Acquisition of that structure was likely the result of a transposition process because it was bracketed by a 5-bp duplication of the target site (CCTCC) that might be likely considered as the signature of transposition. Thus, this genetic structure defines a mobile insertion cassette (mic) containing the qnrS2 gene instead of a transposase, as observed in insertion sequences. Those mic elements are not related to the gene cassette associated with class 1 integrons (26). The mic-qnrS2 element identified here carried putative promoter sequences able to enhance the qnrS2 expression made of a –35 box (TTCTCT) and a –10 box (TAACTT) separated by a 17-bp sequence.
Our study identified plasmid-mediated quinolone resistance QnrS determinants from water samples collected in different sites in a Paris river. To our knowledge, this is the first identification of plasmid-mediated QnrS determinants in nonenterobacterial species. Previous studies did not identify such qnr genes from tested gram-negative isolates that represented Campylobacter jejuni (27), Aeromonadaceae, Pseudomonadaceae, Xanthomonadaceae, Moraxellaceae, and Shewanellaceae (11). Identification of QnrS-positive isolates at 2 collection sites in different water samples may highlight their relative persistence in the environment, at least in this area at that time.
The high-level resistance to quinolones and fluoroquinolones might be due to mutations in type II topoisomerase genes because the mutations described in type II topoisomerases in A. media and A. punctata subsp. punctata have already been associated with resistance in Aeromonas spp (18). QnrS2 may confer low-level resistance to quinolones, as known in E. coli.
The qnrS1 gene has been identified now from several enterobacterial isolates from Japan (9), Germany (28), the United Kingdom (29), the United States (25), France (17), Vietnam (30), Taiwan (31,32), and Denmark (33). The qnrS2 gene (92% amino acid identity with QnrS1) was identified from a transferable IncQ-related plasmid (pGNB2) isolated from an activated sludge bacterial community of a wastewater treatment plant in Germany (24) and in a single non-Typhi Salmonella clinical isolate from the United States (25). Identification of QnrS determinants in Aeromonas spp. indicates that those bacterial species may play a role as a reservoir of the qnrS genes in an aquatic environment, as already evidenced for tet genes (34,35). However, whether Aeromonas species are a main or an accessory reservoir of plasmid-mediated quinolone resistance determinants in regard to Enterobacteriaceae remains to be determined. For Aeromonas spp. to act as a reservoir of qnr genes it must be capable of acquiring these resistance genes from their progenitors (36) and transferring this genetic information to Enterobacteriaceae. QnrS2-positive plasmids p37 and p42 were not able to be transferred by conjugation to an E. coli host in vitro, but they were able to replicate in E. coli, indicating their broad host spectrum. In addition, our study demonstrated that this IncU-type plasmid-mediated qnrS2 gene was expressed and able to confer reduced susceptibility to quinolones, at least in E. coli. Aeromonas species and IncU plasmids, which are ubiquitous in a wide range of environments, might therefore act as important vectors for transfer of plasmid-mediated quinolone resistance determinants (23).
As opposed to most qnrA and qnrB genes, qnrS genes have never been reported to be associated with sul1-type class 1 integrons (6,8). The qnrS1 gene has been identified either upstream of Tn3-like transposon (9,28) or upstream of the insertion sequence ISEcl2 (17,37). In IncQ-related plasmid pGNB2 and in pMG308 from a Salmonella isolate, surrounding genetic structures of the qnrS2 genes were similar, with 2 open reading frames located immediately downstream of qnrS2, similar to repC and repA genes involved in plasmid replication (24,25) (Figure 2). In plasmid p37 from A. punctata 37, the genetic structure was different since the qnrS2 was part of a transposon-like structure and inserted in an open reading frame coding for a zinc MpR.
We have shown that plasmid integration of the qnrS2 gene may result from a peculiar transposition process that likely corresponds to trans-transposition. The qnrS2 gene was inserted in a peculiar mic. Such mic elements have been identified rarely, e.g., mic231-like elements in Bacillus cereus, carrying in only 1 instance an antibiotic resistance gene, the fos gene encoding resistance to fosfomycin (26). This finding may indicate that mic elements might be clinically relevant and the origin of an additional gene plasticity in a bacterial species. These elements containing genetic features involved in gene dissemination (with a transposase likely acting in-trans) and expression may be also vehicles for antibiotic resistance genes. This structure type may be added to the list of genetic tools at the origin of dissemination and expression of antibiotic resistance determinants.
As previously described for the spread of a carbapenemase gene (blaIMI-2) in US rivers (20), this report underlines that the aquatic environment is an important reservoir of novel antibiotic resistance determinants. Quinolones are antimicrobial agents extensively used in aquaculture and are stable molecules in water (as opposed to β-lactams) (38). Thus, they may be the source of an important driving force for selection of quinolone resistance, which explains why QnrS2-positive plasmids did not possess any additional resistance determinants. Further studies might focus on the particular effect of quinolone use for inducing the qnrS2 gene mobility because it is known those molecules may induce bacterial repair systems and antibiotic resistance gene transfer (39).
We have previously shown that the qnrA and qnrS genes originate from water-borne bacterial species, S. algae and Vibrio splendidus, respectively. This identification of a qnrS gene in another water-borne species, Aeromonas, further strenghens the role of water as a vehicle for spread of those resistance determinants.
Dr Cattoir is clinical microbiologist at the Hospital Henri Mondor in Créteil and a PhD student at the Hôpital de Bicêtre, Institut Nationale de la Santé et de la Recherche Médicale Unité 914, South-Paris Medical School, University Paris XI, France. His research focuses on emerging resistance mechanisms to quinolone molecules in clinical and environmental bacterial species.
Acknowledgment
This work was funded by a grant from the Ministère de l’Education Nationale et de la Recherche (UPRES-EA3539), Université Paris XI, France, and primarily by a grant from the European Community (6th PCRD, LSHM-CT-2005-018705).
References
- Hooper DC. Emerging mechanisms of fluoroquinolone resistance. Emerg Infect Dis. 2001;7:337–41.PubMedGoogle Scholar
- Ruiz J. Mechanisms of resistance to quinolones: target alterations, decreased accumulation and DNA gyrase protection. J Antimicrob Chemother. 2003;51:1109–17. DOIPubMedGoogle Scholar
- Martinez-Martinez L, Pascual A, Jacoby GA. Quinolone resistance from a transferable plasmid. Lancet. 1998;351:797–9. DOIPubMedGoogle Scholar
- Tran JH, Jacoby GA, Hooper DC. Interaction of the plasmid-encoded quinolone resistance protein Qnr with Escherichia coli DNA gyrase. Antimicrob Agents Chemother. 2005;49:118–25. DOIPubMedGoogle Scholar
- Tran JH, Jacoby GA, Hooper DC. Interaction of the plasmid-encoded quinolone resistance protein QnrA with Escherichia coli topoisomerase IV. Antimicrob Agents Chemother. 2005;49:3050–2. DOIPubMedGoogle Scholar
- Nordmann P, Poirel L. Emergence of plasmid-mediated resistance to quinolones in Enterobacteriaceae. J Antimicrob Chemother. 2005;56:463–9. DOIPubMedGoogle Scholar
- Martinez-Martinez L, Pascual A, Garcia I, Tran J, Jacoby GA. Interaction of plasmid and host quinolone resistance. J Antimicrob Chemother. 2003;51:1037–9. DOIPubMedGoogle Scholar
- Robicsek A, Jacoby GA, Hooper DC. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect Dis. 2006;6:629–40. DOIPubMedGoogle Scholar
- Hata M, Suzuki M, Matsumoto M, Takahashi M, Sato K, Ibe S, Cloning of a novel gene for quinolone resistance from a transferable plasmid in Shigella flexneri 2b. Antimicrob Agents Chemother. 2005;49:801–3. DOIPubMedGoogle Scholar
- Jacoby GA, Walsh KE, Mills DM, Walker VJ, Oh H, Robicsek A, qnrB, another plasmid-mediated gene for quinolone resistance. Antimicrob Agents Chemother. 2006;50:1178–82. DOIPubMedGoogle Scholar
- Poirel L, Rodriguez-Martinez JM, Mammeri H, Liard A, Nordmann P. Origin of plasmid-mediated quinolone resistance determinant QnrA. Antimicrob Agents Chemother. 2005;49:3523–5. DOIPubMedGoogle Scholar
- Cattoir V, Poirel L, Mazel D, Soussy CJ, Nordmann P. Vibrio splendidus as the source of plasmid-mediated QnrS-like quinolone resistance determinants. Antimicrob Agents Chemother. 2007;51:2650–1. DOIPubMedGoogle Scholar
- Poirel L, Liard A, Rodriguez-Martinez JM, Nordmann P. Vibrionaceae as a possible source of Qnr-like quinolone resistance determinants. J Antimicrob Chemother. 2005;56:1118–21. DOIPubMedGoogle Scholar
- Dortet L, Legrand P, Soussy CJ, Cattoir V. Bacterial identification, clinical significance, and antimicrobial susceptibilities of Acinetobacter ursingii and Acinetobacter schindleri, two frequently misidentified opportunistic pathogens. J Clin Microbiol. 2006;44:4471–8. DOIPubMedGoogle Scholar
- Yanez MA, Catalan V, Apraiz D, Figueras MJ, Martinez-Murcia AJ. Phylogenetic analysis of members of the genus Aeromonas based on gyrB gene sequences. Int J Syst Evol Microbiol. 2003;53:875–83. DOIPubMedGoogle Scholar
- Cattoir V, Poirel L, Rotimi V, Soussy CJ, Nordmann P. Multiplex PCR for detection of plasmid-mediated quinolone resistance qnr genes in ESBL-producing enterobacterial isolates. J Antimicrob Chemother. 2007;60:394–7. DOIPubMedGoogle Scholar
- Poirel L, Leviandier C, Nordmann P. Prevalence and genetic analysis of plasmid-mediated quinolone resistance determinants QnrA and QnrS in Enterobacteriaceae isolates from a French university hospital. Antimicrob Agents Chemother. 2006;50:3992–7. DOIPubMedGoogle Scholar
- Goni-Urriza M, Arpin C, Capdepuy M, Dubois V, Caumette P, Quentin C. Type II topoisomerase quinolone resistance-determining regions of Aeromonas caviae, A. hydrophila, and A. sobria complexes and mutations associated with quinolone resistance. Antimicrob Agents Chemother. 2002;46:350–9. DOIPubMedGoogle Scholar
- Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; 17th informational supplement M100–S17. Wayne (PA): The Institute; 2007.
- Aubron C, Poirel L, Ash RJ, Nordmann P. Carbapenemase-producing Enterobacteriaceae, US rivers. Emerg Infect Dis. 2005;11:260–4.PubMedGoogle Scholar
- Casas C, Anderson EC, Ojo KK, Keith I, Whelan D, Rainnie D, Characterization of pRAS1-like plasmids from atypical North American psychrophilic Aeromonas salmonicida. FEMS Microbiol Lett. 2005;242:59–63. DOIPubMedGoogle Scholar
- Kieser T. Factors affecting the isolation of CCC DNA from Streptomyces lividans and Escherichia coli. Plasmid. 1984;12:19–36. DOIPubMedGoogle Scholar
- Rhodes G, Parkhill J, Bird C, Ambrose K, Jones MC, Huys G, Complete nucleotide sequence of the conjugative tetracycline resistance plasmid pFBAOT6, a member of a group of IncU plasmids with global ubiquity. Appl Environ Microbiol. 2004;70:7497–510. DOIPubMedGoogle Scholar
- Bonemann G, Stiens M, Puhler A, Schlueter A. Mobilizable IncQ-related plasmid carrying a new quinolone resistance gene, qnrS2, isolated from the bacterial community of a wastewater treatment plant. Antimicrob Agents Chemother. 2006;50:3075–80. DOIPubMedGoogle Scholar
- Gay K, Robicsek A, Strahilevitz J, Park CH, Jacoby GA, Barrett TJ, Plasmid-mediated quinolone resistance in non-Typhi serotypes of Salmonella enterica. Clin Infect Dis. 2006;43:297–304. DOIPubMedGoogle Scholar
- De Palmenaer D, Vermeiren C, Mahillon J. IS231-MIC231 elements from Bacillus cereus sensu lato are modular. Mol Microbiol. 2004;53:457–67. DOIPubMedGoogle Scholar
- Chatzipanagiotou S, Ioannidou V, Ioannidis A, Nicolaou C, Papavasileiou E, Chaniotaki S, Absence of the plasmid-mediated quinolone resistance qnrA gene among Campylobacter jejuni clinical isolates from Greece. Int J Antimicrob Agents. 2005;26:261–2. DOIPubMedGoogle Scholar
- Kehrenberg C, Friederichs S, de Jong A, Michael GB, Schwarz S. Identification of the plasmid-borne quinolone resistance gene qnrS in Salmonella enterica serovar Infantis. J Antimicrob Chemother. 2006;58:18–22. DOIPubMedGoogle Scholar
- Hopkins KL, Wootton L, Day MR, Threlfall EJ. Plasmid-mediated quinolone resistance determinant qnrS1 found in Salmonella enterica strains isolated in the UK. J Antimicrob Chemother. 2007;59:1071–5. DOIPubMedGoogle Scholar
- Poirel L, Nguyen TV, Weintraub A, Leviandier C, Nordmann P. Plasmid-mediated quinolone resistance determinant qnrS in Enterobacter cloacae. Clin Microbiol Infect. 2006;12:1021–3. DOIPubMedGoogle Scholar
- Chen YT, Shu HY, Li LH, Liao TL, Wu KM, Shiau YR, Complete nucleotide sequence of pK245, a 98-kilobase plasmid conferring quinolone resistance and extended-spectrum-beta-lactamase activity in a clinical Klebsiella pneumoniae isolate. Antimicrob Agents Chemother. 2006;50:3861–6. DOIPubMedGoogle Scholar
- Wu JJ, Ko WC, Tsai SH, Yan JJ. Prevalence of plasmid-mediated quinolone resistance determinants QnrA, QnrB, and QnrS among clinical isolates of Enterobacter cloacae in a Taiwanese hospital. Antimicrob Agents Chemother. 2007;51:1223–7. DOIPubMedGoogle Scholar
- Cavaco LM, Hansen DS, Friis-Moller A, Aarestrup FM, Hasman H, Frimodt-Moller N. First detection of plasmid-mediated quinolone resistance (qnrA and qnrS) in Escherichia coli strains isolated from humans in Scandinavia. J Antimicrob Chemother. 2007;59:804–5. DOIPubMedGoogle Scholar
- Rhodes G, Huys G, Swings J, McGann P, Hiney M, Smith P, Distribution of oxytetracycline resistance plasmids between aeromonads in hospital and aquaculture environments: implication of Tn1721 in dissemination of the tetracycline resistance determinant tetA. Appl Environ Microbiol. 2000;66:3883–90. DOIPubMedGoogle Scholar
- Schmidt AS, Bruun MS, Dalsgaard I, Larsen JL. Incidence, distribution, and spread of tetracycline resistance determinants and integron-associated antibiotic resistance genes among motile aeromonads from a fish farming environment. Appl Environ Microbiol. 2001;67:5675–82. DOIPubMedGoogle Scholar
- Young HK. Antimicrobial resistance spread in aquatic environments. J Antimicrob Chemother. 1993;31:627–35. DOIPubMedGoogle Scholar
- Poirel L, Cattoir V, Soares A, Soussy CJ, Nordmann P. Novel Ambler class A β-lactamase LAP-1 and its association with the plasmid-mediated quinolone resistance determinant QnrS1. Antimicrob Agents Chemother. 2006;51:631–7. DOIPubMedGoogle Scholar
- Kümmerer K. Resistance in the environment. J Antimicrob Chemother. 2004;54:311–20. DOIPubMedGoogle Scholar
- Beaber JW, Hochhut B, Waldor MK. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature. 2004;427:72–4. DOIPubMedGoogle Scholar
Figures
Table
Cite This ArticleTable of Contents – Volume 14, Number 2—February 2008
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Patrice Nordmann, Service de Bactériologie-Virologie-Hygiène, Institut Nationale de la Santé et de la Recherche Médicale Unité 914, Hôpital de Bicêtre, 78 rue du Général Leclerc, 94275 Le Kremlin-Bicêtre, France;
Top