Volume 14, Number 9—September 2008
Dispatch
Juquitiba-like Hantavirus from 2 Nonrelated Rodent Species, Uruguay
Figure 2
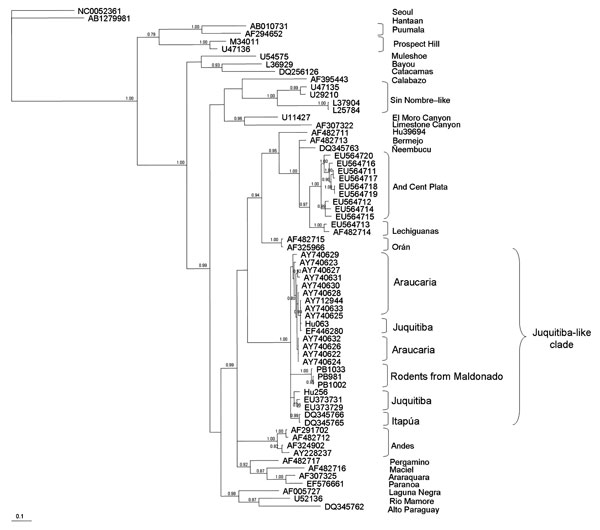
Figure 2. Majority-rule consensus tree obtained in the Bayesian analysis of sequences of the small segment of Juquitiba-like hantavirus isolated from 2 nonrelated rodent species. Posterior probabilities >0.80 are shown at the nodes. Analyses were performed as described in Figure 1. Seropositive specimens from Uruguay are as follows: PB1033 (black-footed pigmy rice rat, Oligoryzomys nigripes), PB981 and PB1002 (long-nosed mouse, Oxymycterus nasutus), GenBank accession nos. EU564724, EU564722, EU564723, respectively. Analyzed hantavirus sequences are Seoul (NC0052361), Hantaan (AB1279981), Laguna Negra (AF005727), Rio Mamore (U52136), Alto Paraguay (DQ345762), Andes Central Plata (EU564711, EU564717, EU564718, EU564719, EU564716, EU564720, EU564715), Lechiguanas (EU564713, AF482714), Bermejo (AF482713), Ñeembucu (DQ345763), Hu39694 (AF482711), Orán (AF482715, AF325966), Araucaria (AY740627, AY712944, AY740631, AY740633, AY740625, AY740628, AY740632, AY7406261, AY7406221, AY7406241, AY740630, AY740629, AY740623), Juquitiba (Hu063, EF446280, Hu256, EU373729, EU373731), Itapúa (DQ345766, DQ345765), Andes (AF291702, AF482712, AF324902, AY228237), Maciel (AF482716), Araraquara (AF307325), Paranoa (EF576661), Pergamino (AF482717), Sin Nombre–like (U47135, U29210, L37904, L25784), Calabazo (AF395443); El Moro Canyon (U11427), Limestone Canyon (AF307322), Bayou (L36929), Catacamas (DQ256126), Muleshoe (U54575), Puumala (AB010731, AF294652), and Prospect Hill (M34011, U47136). Scale bar indicates expected changes per site.