Volume 15, Number 4—April 2009
Dispatch
Isolation of Genotype V St. Louis Encephalitis Virus in Florida
Appendix Figure
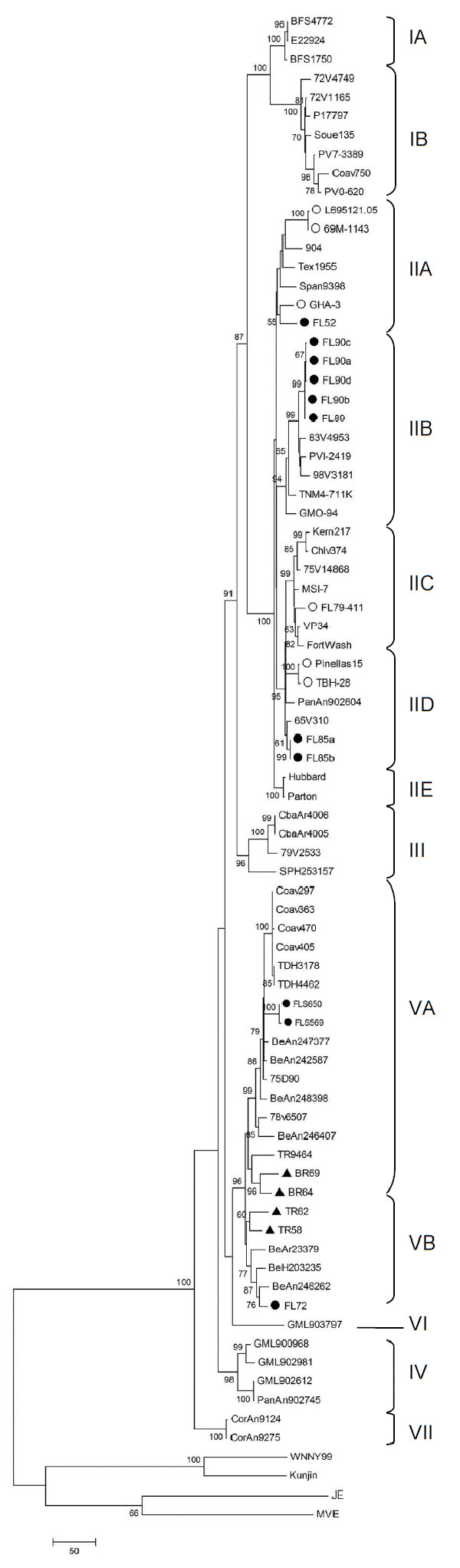
Appendix Figure. Phylogram of the complete envelope region of St. Louis encephalitis virus (SLEV) strains, inferred using the maximum parsimony method in MEGA4 software (14). Bootstrap analysis was performed using 1,000 replicates, and the consensus tree (generated by majority rule of 27 most parsimonious trees) was chosen. The number at each node indicates percent branch support by bootstrap sampling; values <50 were collapsed. Branch lengths represent the amount of genetic divergence; the scale bar corresponds to number of base changes in the sequence. The phylogram includes 11 newly sequenced Florida SLEV strains (●), 6 previously sequenced Florida strains (○) (9), and 4 newly sequenced South American strains (▲). The phylogram also identified 7 lineages, shown as described in an earlier study of 62 strains (9). Florida Genotype V viruses cluster in Lineage VA (FLS569, FLS650) and Lineage VB (FL72).
References
- Lindenbach BD, Rice CM. Flaviviridae: the viruses and their replication. In: Knipe DM, Howley PM, editors. Field’s virology, 4th ed., vol. 1. Philadelphia: Lippincott Williams & Wilkins; 2001. p. 991–1041.
- Burke DS, Monath TP. Flaviviruses. In: Knipe DM, Howley PM, eds. Field’s virology, 4th ed., vol. 1. Philadelphia: Lippincott Williams & Wilkins; 2001. p. 1043–125.
- Fang Y, Reisen WK. Previous infection with West Nile or St. Louis encephalitis viruses provides cross protection during reinfection in house finches. Am J Trop Med Hyg. 2006;75:480–5.PubMedGoogle Scholar
- Reisen WK, Lothrop HD, Wheeler SS, Kennsington M, Gutierrez A, Fang Y, Persistent West Nile virus transmission and the apparent displacement St. Louis encephalitis virus in southeastern California, 2003–2006. J Med Entomol. 2008;45:494–508. DOIPubMedGoogle Scholar
- Bigler BS. St. Louis encephalitis. 1999 [cited 2008 Jul 1]. Available from http://www.doh.state.fl.us/disease_ctrl/epi/htopics/reports/slepres2.pdf
- Nelson DB, Kappus KD, Janowski HT, Buff E, Wellings FM, Schneider NJ. St. Louis encephalitis—Florida 1977. Patterns of a widespread outbreak. Am J Trop Med Hyg. 1983;32:412–6.PubMedGoogle Scholar
- Day JF. Predicting St. Louis encephalitis virus epidemics: lessons from recent, and not so recent, outbreaks. Annu Rev Entomol. 2001;46:111–38. DOIPubMedGoogle Scholar
- Reisen WK, Presser SB, Lin J, Enge B, Hardy JL, Emmons RW. Viremia and serological responses in adult chickens infected with western equine encephalomyelitis and St. Louis encephalitis viruses. J Am Mosq Control Assoc. 1994;10:549–55.PubMedGoogle Scholar
- Kramer LD, Chandler LJ. Phylogenic analysis of the envelope gene of St. Louis encephalitis virus. Arch Virol. 2001;146:2341–55. DOIPubMedGoogle Scholar
- Florida Department of Health. Arbovirus surveillance: annual and laboratory reports, 1999–2007 [cited 2008 Jul 1]. Available from http://www.doh.state.fl.us/ENVIRONMENT/community/arboviral/survey-info.htm
- Blackmore CG, Stark LM, Jeter WC, Oliveri RL, Brooks RG, Conti LA, Surveillance results from the first West Nile virus transmission season in Florida, 2001. Am J Trop Med Hyg. 2003;69:141–50.PubMedGoogle Scholar
- Ciota AT, Lovelace AO, Ngo KA, Le AN, Maffei JG, Franke MA, Cell-specific adaptation of two flaviviruses following serial passage in mosquito cell culture. Virology. 2007;357:165–74. DOIPubMedGoogle Scholar
- Lanciotti RS, Kerst AJ. Nucleic acid sequence-based amplification assays for rapid detection of West Nile and St. Louis encephalitis viruses. J Clin Microbiol. 2001;39:4506–13. DOIPubMedGoogle Scholar
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–9. DOIPubMedGoogle Scholar
- May FJ, Li L, Zhang S, Guzman H, Beasley DW, Tesh RB, Genetic variation of St. Louis encephalitis virus. J Gen Virol. 2008;89:1901–10. DOIPubMedGoogle Scholar