Volume 15, Number 9—September 2009
Research
Distant Relatives of Severe Acute Respiratory Syndrome Coronavirus and Close Relatives of Human Coronavirus 229E in Bats, Ghana
Figure 3
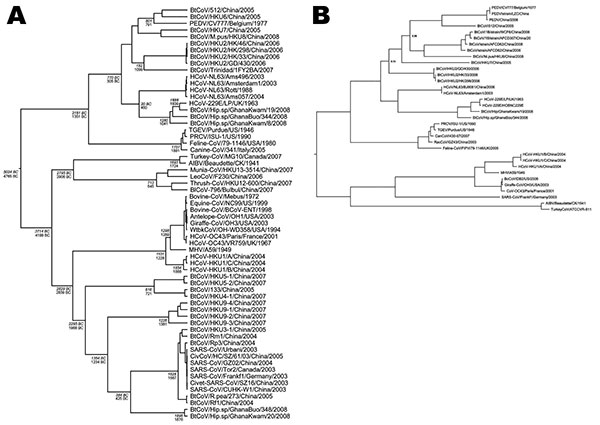
Figure 3. A) Phylogeny of coronaviruses (CoVs) in the RNA-dependent RNA polymerase gene (RdRp, 817-bp fragment) with root point dates derived from Bayesian inference under a relaxed lognormal molecular clock assumption with a codon-based substitution model (SRD06) and an assumption of expansion growth of the virus population. Estimated dates of diversification of CoV lineages at root points are shown in italics for the expansion growth population model and in regular type for the exponential growth model. Dates
Page created: December 07, 2010
Page updated: December 07, 2010
Page reviewed: December 07, 2010
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.