Volume 16, Number 8—August 2010
Research
Bat Coronaviruses and Experimental Infection of Bats, the Philippines
Figure 1
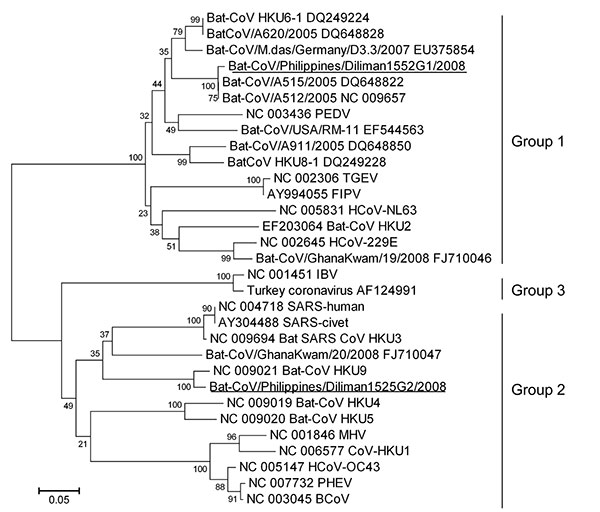
Figure 1. Phylogenetic tree based on deduced amino acid sequences of partial RNA-dependent RNA polymerase of coronaviruses (CoVs), the Philippines. The 2 new viruses detected in this study are underlined. Percentage of replicate trees in which the associated taxa clustered in the bootstrap test (1,000 replicates) is shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. Evolutionary distances were computed by using the Poisson correction method and are shown as number of amino acid substitutions per site. All positions containing gaps and missing data were eliminated from the dataset. The final dataset included 120 positions. Phylogenetic analyses were conducted in MEGA4 (14). Coronaviruses used for comparisons and their GenBank accession numbers are human coronavirus (HCoV) 229E (HCoV-229E) (NC_002645), porcine epidemic diarrhea virus (PEDV) (NC_003436), transmissible gastroenteritis virus (TGEV) (NC_002306), feline infectious peritonitis virus (FIPV) (AY994055), human coronavirus NL63 (HCoV-NL63) (NC_005831), bat-CoV/A512/2005 (NC_009657), bat-CoV/A515/2005 (DQ648822), bat-CoV/A620/2005 (DQ648828), bat-CoV/A911/2005 (DQ648850), bat-CoV/GhanaKwan/19/2008 (FJ710046), bat-CoV/GhanaKwan/20/2008 (FJ710047), bat-CoV/M.das/Germany/D3.3/2007 (EU375854), bat-CoV/USA/RM-11 (EF544563), bat-CoV HKU2 (EF203064), HKU4 (NC_009019), HKU5 (NC_009020), HKU6 (DQ249224), HKU8 (DQ249228), HKU9 (NC_009021), CoV-HKU1 (NC_006577), human coronavirus (HCoV-OC43) (NC_005147), murine hepatitis virus (MHV) (NC_001846), bovine coronavirus (BCoV) (NC_003045), porcine hemagglutinating encephalomyelitis virus (PHEV) (NC_007732), human severe acute respiratory syndrome coronavirus (SARS) (SARS-human) (NC_004718), civet SARS-like coronavirus (SARS-civet) (AY304488), bat-SARS-like coronavirus HKU3 (bat-SARS-CoV HKU3) (NC_009694), infectious bronchitis virus (IBV) (NC_001451), and turkey coronavirus (AF124991).
References
- Guan Y, Zheng BJ, He YQ, Liu XL, Zhuang ZX, Cheung CL, Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–8. DOIPubMedGoogle Scholar
- Kan B, Wang M, Jing H, Xu H, Jiang X, Yan M, Molecular evolution analysis and geographic investigation of severe acute respiratory syndrome coronavirus-like virus in palm civets at an animal market and on farms. J Virol. 2005;79:11892–900. DOIPubMedGoogle Scholar
- Lau SK, Woo PC, Li KS, Huang Y, Tsoi HW, Wong BH, Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc Natl Acad Sci U S A. 2005;102:14040–5. DOIPubMedGoogle Scholar
- Poon LL, Chu DK, Chan KH, Wong OK, Ellis TM, Leung YH, Identification of a novel coronavirus in bats. J Virol. 2005;79:2001–9. DOIPubMedGoogle Scholar
- Tang XC, Zhang JX, Zhang SY, Wang P, Fan XH, Li LF, Prevalence and genetic diversity of coronaviruses in bats from China. J Virol. 2006;80:7481–90. DOIPubMedGoogle Scholar
- Woo PC, Lau SK, Li KS, Poon RW, Wong BH, Tsoi HW, Molecular diversity of coronaviruses in bats. Virology. 2006;351:180–7. DOIPubMedGoogle Scholar
- Vijaykrishna D, Smith GJ, Zhang JX, Peiris JS, Chen H, Guan Y. Evolutionary insights into the ecology of coronaviruses. J Virol. 2007;81:4012–20. DOIPubMedGoogle Scholar
- Carrington CV, Foster JE, Zhu HC, Zhang JX, Smith GJ, Thompson N, Detection and phylogenetic analysis of group 1 coronaviruses in South American bats. Emerg Infect Dis. 2008;14:1890–3. DOIPubMedGoogle Scholar
- Dominguez SR, O'Shea TJ, Oko LM, Holmes KV. Detection of group 1 coronaviruses in bats in North America. Emerg Infect Dis. 2007;13:1295–300.PubMedGoogle Scholar
- Gloza-Rausch F, Ipsen A, Seebens A, Gottsche M, Panning M, Felix Drexler J, Detection and prevalence patterns of group I coronaviruses in bats, northern Germany. Emerg Infect Dis. 2008;14:626–31. DOIPubMedGoogle Scholar
- Misra V, Dumonceaux T, Dubois J, Willis C, Nadin-Davis S, Severini A, Detection of polyoma and corona viruses in bats of Canada. J Gen Virol. 2009;90:2015–22. DOIPubMedGoogle Scholar
- Pfefferle S, Oppong S, Drexler JF, Gloza-Rausch F, Ipsen A, Seebens A, Distant relatives of severe acute respiratory syndrome coronavirus and close relatives of human coronavirus 229E in bats, Ghana. Emerg Infect Dis. 2009;15:1377–84. DOIPubMedGoogle Scholar
- de Souza Luna LK, Heiser V, Regamey N, Panning M, Drexler JF, Mulangu S, Generic detection of coronaviruses and differentiation at the prototype strain level by reverse transcription–PCR and nonfluorescent low-density microarray. J Clin Microbiol. 2007;45:1049–52. DOIPubMedGoogle Scholar
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–9. DOIPubMedGoogle Scholar
- Baric RS, Shieh CK, Stohlman SA, Lai MM. Analysis of intracellular small RNAs of mouse hepatitis virus: evidence for discontinuous transcription. Virology. 1987;156:342–54. DOIPubMedGoogle Scholar
- Hiscox JA, Mawditt KL, Cavanagh D, Britton P. Investigation of the control of coronavirus subgenomic mRNA transcription by using T7-generated negative-sense RNA transcripts. J Virol. 1995;69:6219–27.PubMedGoogle Scholar
- Woo PC, Wang M, Lau SK, Xu H, Poon RW, Guo R, Comparative analysis of twelve genomes of three novel group 2c and group 2d coronaviruses reveals unique group and subgroup features. J Virol. 2007;81:1574–85. DOIPubMedGoogle Scholar
- Simmons N. Order Chiroptera. In: Wilson DE, Reeder DM, editors. Mammal species of the world: a taxonomic and geographic reference. Baltimore: The Johns Hopkins University Press; 2005. p. 312–529.