Volume 5, Number 1—February 1999
Research
Genetic Diversity and Distribution of Peromyscus-Borne Hantaviruses in North America
Figure 1
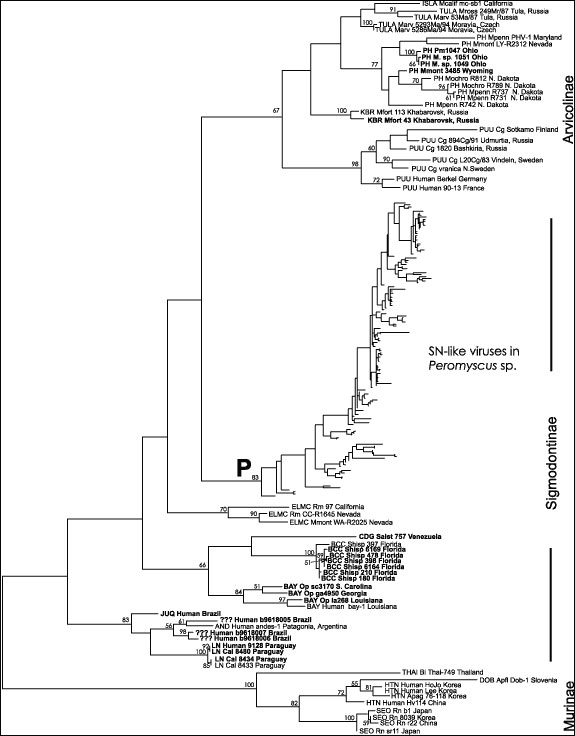
Figure 1. Overall hantavirus phylogenetic tree based on analysis of a 139 nucleotide fragment of the G2 coding region of the virus M segment. All newly reported sequences are shown bolded. The three virus groups corresponding to the hantaviruses carried by rodents of the subfamilies Murinae, Sigmodontinae and Arvicolinae are indicated. P indicates the clade containing the Sin Nombre- like viruses found in Peromyscus species rodents, the details of which are shown in Figure 2. Horizontal branch lengths are proportional to the nucleotide step differences between taxa and predicted nodes. No scale bar is indicated as the actual number values are arbitary due to the weighting used in the successive approximations method (see Appendix for details). Bootstrap values greater than 50% are indicated above branches. Virus labels include the virus or virus lineage name (ISLA, Isla Vista; TULA, Tula; PH, Prospect Hill or Prospect Hill-like; KBR, Khabarovsk; PUU, Puumala; SN, Sin Nombre; ELMC, El Moro Canyon; CDG, Caño Delgadito; BCC, Black Creek Canal; BAY, Bayou; JUQ, Juquitiba; AND, Andes; LN, Laguna Negra; THAI, Thailand; DOB, Dobrava; HTN, Hantaan; SEO, Seoul), species source of material (Mcalif, Microtus californicus; Mross, Microtus rossiaemeridionalis; Marv, Microtus arvalis; Mpenn, Microtus pennsylvanicus; Mmont, Microtus montanus, Mochro, Microtus ochrogaster; Mfort, Microtus fortis; Cg, Clethrionomys glareolus; Rm, Reithrodontomys megalotis; Salst, Sigmodon alstoni; Shisp, Sigmodon hispidus; Op, Oryzomys palustris; Cal, Calomys laucha; Bi, Bandicota indica; Apfl, Apodemus flavicollis; Apag, Apodemus agrarius; Rn Rattus norvegicus), identifier, and state, region or country of origin. For spreadsheet containing the details of all samples, see Technical Appendix 1.