Volume 13, Number 6—June 2007
Research
Antimicrobial Drug–Resistant Escherichia coli from Humans and Poultry Products, Minnesota and Wisconsin, 2002–2004
Cite This Article
Citation for Media
Abstract
The food supply, including poultry products, may transmit antimicrobial drug–resistant Escherichia coli to humans. To assess this hypothesis, 931 geographically and temporally matched E. coli isolates from human volunteers (hospital inpatients and healthy vegetarians) and commercial poultry products (conventionally raised or raised without antimicrobial drugs) were tested by PCR for phylogenetic group (A, B1, B2, D) and 60 virulence genes associated with extraintestinal pathogenic E. coli. Isolates resistant to trimethoprim-sulfamethoxazole, quinolones, and extended-spectrum cephalosporins (n = 331) were compared with drug-susceptible isolates (n = 600) stratified by source. Phylogenetic and virulence markers of drug-susceptible human isolates differed considerably from those of human and poultry isolates. In contrast, drug-resistant human isolates were similar to poultry isolates, and drug-susceptible and drug-resistant poultry isolates were largely indistinguishable. Many drug-resistant human fecal E. coli isolates may originate from poultry, whereas drug-resistant poultry-source E. coli isolates likely originate from susceptible poultry-source precursors.
Acquired resistance to first-line antimicrobial agents increasingly complicates the management of extraintestinal infections due to Escherichia coli, which are a major source of illness, death, and increased healthcare costs (1–4). One suspected source of drug-resistant E. coli in humans is use of antimicrobial drugs in agriculture. This use presumably selects for drug-resistant E. coli, which may be transmitted to humans through the food supply (5–7). Supporting this hypothesis is the high prevalence of antimicrobial drug–resistant E. coli in retail meat products, especially poultry (8–11), and the similar molecular characteristics of fluoroquinolone-resistant E. coli from chicken carcasses and from colonized and infected persons in Barcelona, Spain, in contrast to the marked differences between drug-susceptible and drug-resistant source isolates from humans (12).
To further assess the poultry-human connection, we used molecular typing to characterize drug-resistant and drug-susceptible E. coli isolates from feces of human volunteers or newly hospitalized patients in Minnesota and Wisconsin and from poultry products sold or processed in the same region. Resistance phenotypes of interest include trimethoprim-sulfamethoxazole (TMP-SMZ), quinolones/fluoroquinolones, and extended-spectrum cephalosporins. These agents are used for treatment of human E. coli infections. These drugs (or congeners) are also used in poultry production (e.g., each year in the United States an estimated 1.6 billion broiler eggs or chicks receive ceftiofur [13]); E. coli isolates resistant to these drugs are found in poultry. We examined according to phylogenetic group distribution and virulence gene profile, whether drug-resistant human isolates more closely resemble susceptible human isolates, which is consistent with acquisition of resistance within humans, or instead resemble poultry isolates, which is consistent with foodborne transmission of poultry-source organisms to humans. We also examined whether poultry-source resistant and susceptible isolates are similar, which is consistent with emergence of resistance on farms under selection from agricultural use of antimicrobial drugs.
Participants and Bacterial Strains
Human fecal samples were collected from 622 adults newly admitted to local hospitals in 4 rural communities in Minnesota (Willmar) or Wisconsin (Eau Claire, La Crosse, and Marshfield) and from 100 healthy self-identified vegetarians in these and nearby communities (14). Hospital patients were recruited from June 2002 through May 2003, vegetarians during the first 6 months of 2004. Fecal samples were collected by study personnel by using rectal swabs (hospital patients) or by the participants (vegetarians). To prevent isolation of hospital-acquired flora, inpatients samples were collected within 36 hours of hospital admission. Guidelines of the authors’ institutions regarding use of human subjects were followed in this study. The relevant institutional review boards reviewed and approved the protocol. All participants provided informed consent.
A total of 180 retail poultry products (155 chicken and 25 turkey) were sampled (14). Conventional brands were purchased systematically from all food markets in the 4 primary study communities from May 2002 through May 2003, with 40 retail items obtained per community (total 160 items). These represented at least 18 plants in 11 states. Twenty samples with labels indicating that the poultry were raised naturally or without antibiotics were purchased in or near the study communities in August 2004. Additionally, 40 freshly slaughtered chicken carcasses from local farmers who raised chickens naturally or without antibiotics were obtained during plant inspections by the Minnesota Department of Agriculture from September 2003 through August 2004. The latter 2 groups of chickens, designated “no antibiotics,” were confirmed to have been raised without antibiotics, based on the product label or by contacting the manufacturer or distributor.
Sample Processing
Human fecal samples were suspended and poultry samples and carcasses were massaged in nutrient broth, which was then incubated overnight at 37°C and stored as aliquots at –80°C in glycerol (14). Portions of these frozen stocks were transferred to vancomycin-supplemented (20 mg/L) Luria-Bertani broth. After overnight incubation, these broths were plated directly onto modified Mueller-Hinton (MMH) agar (Amyes medium) (10) with and without ciprofloxacin (4 mg/L) and (separately) nalidixic acid (32 mg/L), and were then incubated overnight. Samples of these Luria-Bertani broths containing vancomycin were placed in MMH broths supplemented individually with TMP-SMZ (4 mg/L TMP plus 76 mg/mL SMZ), cefoxitin (10 mg/L and 32 mg/L), and ceftazidime (10 mg/L and 32 mg/L). After overnight incubation, these broths were plated onto MMH agar plates supplemented with the corresponding agent (same concentrations) for overnight incubation. Colonies resembling E. coli were identified by using the API-20E System (bioMérieux, Marcy-l’Etoile, France).
Susceptibility Testing
At least 1 E. coli colony was randomly selected from each MMH agar plate and tested for disk susceptibility to 24 antimicrobial agents by using Clinical Laboratory Standards Institute (CLSI)–recommended methods, interpretive criteria, and reference strains (15). For isolates resistant to TMP-SMZ, nalidixic acid, or ciprofloxacin, the MIC was determined by Etest (AB-Biodisk, Sona, Sweden) according to the manufacturer's directions. Isolates from cefoxitin- and ceftazidime-supplemented plates underwent broth dilution MIC determinations with cefotaxime and ceftazidime regardless of disk test results. Isolates were classified as resistant to TMP-SMZ if the TMP MIC was >4 mg/L and the SMZ MIC was >76 mg/L, to quinolones if the nalidixic acid MIC was >32 mg/L, to fluoroquinolones if the ciprofloxacin MIC was >4 mg/L, and to extended-spectrum cephalosporins if the MIC to either cefotaxime or ceftazidime was >16 mg/L. The latter threshold corresponds with intermediate susceptibility per CLSI criteria and includes isolates with potentially clinically relevant reduced susceptibility. Because of the small number of isolates within each resistance phenotype, isolates were classified as resistant if they met any of these resistance criteria. Isolates that did not meet any of these resistance criteria were classified as susceptible, even though they may have had reduced susceptibility to other drug classes.
From each sample, 1 colony of each resistance phenotype (TMP-SMZ, quinolones, fluoroquinolones, extended-spectrum cephalosporins) and 1 susceptible isolate, as available, were selected. If multiple isolates from a given sample exhibited similar disk diffusion susceptibility profiles, genomic profiles as generated by using random amplified polymorphic DNA (RAPD) analysis were compared in the same gel (12). One representative of each unique RAPD genotype (as determined by visual inspection) was arbitrarily selected for further analysis.
Phylogenetic Analysis and Virulence Genotyping
All isolates were categorized as to major E. coli phylogenetic group (A, B1, B2, or D) by a multiplex PCR-based assay (16) (Table 1). Genes encoding proven or putative virulence factors of extraintestinal pathogenic E. coli (ExPEC) were detected in a sequential fashion. All isolates were screened for 5 ExPEC-defining virulence genes and hlyD (hemolysin). Isolates were operationally defined as ExPEC if ≥2 of the following were present: papA and/or papC (P fimbriae structural subunit and assembly), sfa/focDE (S and F1C fimbriae), afa/draBC (Dr binding adhesins), iutA (aerobactin system), and kpsM II (group 2 capsule) (8). All ExPEC isolates were then tested for 60 ExPEC-associated virulence genes and alleles thereof. Testing was conducted by using 2 independently prepared lysates of each isolate and established PCR-based methods (12,17). Isolates from various source groups (e.g., hospital volunteers, conventionally raised poultry) were tested in parallel to avoid cohort effects. The virulence score was the number of virulence genes detected adjusted for multiple detection of the pap, sfa/foc, and kps operons (12).
Statistical Methods
The unit of analysis was the individual isolate. Comparisons of proportions were tested by using Fisher exact test (2-tailed). Comparisons of virulence scores were tested by using Mann-Whitney U test (2-tailed exact probability). Principal coordinates analysis (PCA), also known as metric multidimensional scaling, is a multivariate statistical technique used to provide a simpler, low-dimensional graphic summary of the similarity between multiple samples (e.g., isolates) across multiple loci (18). New axes for plotting the isolates are derived from a data matrix of estimated dissimilarities between isolates. The first 2 principal coordinates, which account for the most variance, are used to plot the data. The distances between points in the plot represent isolate similarity. The dimensions represented by the (statistically uncorrelated) axes have no intrinsic meaning, i.e., they have no units. Using GenAlEx6 (19), we applied PCA to the screening dataset (all isolates) and the extended virulence profile dataset (ExPEC isolates) as a way to collapse the multiple variables for simplified among-group comparisons. For each PCA, results for each isolate from the first 2 PCA axes were used in multiple analysis of variance (MANOVA) to test for among-group differences. These values also were plotted to spatially represent the degree of separation or overlap of isolates on the 2-axis plane. For the ExPEC isolates, pairwise similarity relationships according to extended virulence profiles and phylogenetic group were used to construct a dendrogram according to the unweighted pair group method with arithmetic averages (20). The criterion for statistical significance throughout was p≤0.01 to account for multiple comparisons.
Isolation of Drug-Resistant and Drug-Susceptible E. coli
Selective processing of 942 human fecal and poultry samples yielded 931 unique E. coli isolates, which constituted the study population. Of the 931 isolates, 530 (57%) were from human volunteers and 401 (43%) from poultry products. Of the human isolates, 456 (86%) were from hospital patients and 74 (14%) from vegetarians. Of the poultry isolates, 289 (72%) were from conventionally raised retail poultry and 112 (28%) from poultry raised without antibiotics. The median number of unique E. coli isolates per sample was 1 for human fecal samples and 2 for poultry (range 1–4 for both).
Overall, 331 isolates (70 human, 261 poultry) were classified as resistant on the basis of reduced susceptibility to TMP-SMZ, quinolones/fluoroquinolones, and extended-spectrum cephalosporins. The remaining 600 isolates (460 human, 140 poultry) were susceptible to all these drug classes and were classified as susceptible (regardless of other possible drug resistance). The resistant isolates were distributed by resistance phenotype as follows: TMP-SMZ, 154 (47 human, 107 poultry); quinolones, 115 (26 human, 89 poultry); and extended-spectrum cephalosporins, 114 (14 human, 100 poultry). The 7 fluoroquinolone-resistant isolates (5 human, 2 poultry) were analyzed within the quinolone-resistant group.
Phylogenetic Distribution and Prevalence of ExPEC-defining Markers
The initial screening showed the 931 isolates to be fairly evenly distributed among the 4 major E. coli phylogenetic groups (20%–28% per group). However, they had various prevalences (2%–39% each) of the screening ExPEC virulence genes (Table 1). Overall, 27% of the isolates qualified as ExPEC by having ≥2 of the 5 ExPEC-defining markers (Table 1).
For enhanced resolution of similarities and differences, the 243 available ExPEC isolates underwent extended virulence genotyping for 60 ExPEC-associated virulence genes. All but 6 of these traits were detected in ≥1 isolate each, with prevalences ranging from 0.4% to 98% (Table 2).
Prevalence Comparisons
Phylogenetic group distribution and virulence gene prevalence differed considerably according to source (human versus poultry) and resistance status. This finding is shown in Table 1 for all 931 isolates (screening virulence genes only) and in Table 2 for the 243 ExPEC isolates (extended virulence profiles). Drug-resistant and drug-susceptible human isolates were separately compared with the combined group of all poultry isolates (i.e., all susceptible and resistant). We analyzed poultry isolates as a single group because the distribution of traits was similar in drug-resistant and susceptible poultry isolates; i.e., only 1 trait (iutA) was significantly associated with resistance among poultry isolates.
Consistent differences in phylogenetic and virulence gene distribution were evident between groups (Tables 1, 2). First, drug-susceptible human isolates differed considerably from drug-resistant human isolates. Second, drug-susceptible human isolates differed from poultry isolates. Third, although human drug-resistant isolates and poultry isolates exhibited some differences, these were considerably fewer and less extreme than those between drug-susceptible human isolates and poultry isolates. Similar results were obtained in subgroup analyses when isolates from hospital patient fecal samples were compared separately with isolates from conventionally raised poultry or when isolates from fecal samples from vegetarians were compared separately with isolates from poultry raised without antibiotics.
PCA
PCA was used to concurrently analyze multiple bacterial characteristics. The first PCA was conducted for the total population (n = 931) with the 7 screening virulence genes plus phylogenetic group. According to a 2 × 2 (source × resistance status) MANOVA of the first 2 axes of the PCA (which accounted for 65% of total variance), all 3 independent variables considered (source, resistance status, and interaction term) showed a p value ≤0.001. Accordingly, pairwise comparisons were made between individual source-resistance groups by 1-factor MANOVA. Susceptible human isolates differed (p<0.001) from each of the other 3 groups, whereas the other 3 groups differed marginally from each other. The individual axes supported this conclusion. These axes showed more extreme differences between drug-susceptible human isolates and each of the other 3 groups (p<0.001 for 5 of 6 comparisons) than among the other groups (p>0.01 for 4 of 6 comparisons).
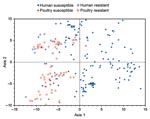
Next, PCA was conducted for the 243 available ExPEC isolates based on all 60 virulence genes plus phylogenetic group. According to an initial 2 × 2 MANOVA of the results from the first 2 PCA axes (which accounted for 57% of total variance), all 3 independent variables (source, resistance status, and interaction term) showed a p value <0.001. Accordingly, pairwise comparisons were made between individual source-resistance groups by 1-factor MANOVA. Susceptible human isolates differed (p<0.001) from each of the other 3 groups, whereas the other 3 groups did not differ significantly from each another. In a plot of the (axis 1–axis 2) plane, drug-susceptible poultry isolates, drug-resistant poultry isolates, and drug-resistant human isolates overlapped and were confined largely to the left half of the grid (negative values on axis 1). In contrast, drug-susceptible human isolates, although overlapping somewhat with these groups, were concentrated principally within the right half of the grid (positive values on axis 1) (Figure 1).
Aggregate Virulence Scores
The various source and resistance groups were also compared for aggregate virulence scores (ExPEC isolates only). According to virulence score distribution, drug-susceptible human isolates (higher scores) segregated widely from the other 3 subgroups (lower scores), which were largely superimposed on each another (Figure 2). Because drug-resistant and drug-susceptible poultry isolates had similar virulence scores, they were combined for statistical analysis. Drug-susceptible human isolates had the highest scores (median 13.0, range 4.25–20.0). Drug-resistant human and poultry isolates had significantly lower scores that did not differ between humans and poultry (median 9.0, range 6.0–15.25, and median 8.75, range 3.75–15.0, respectively; vs. drug-susceptible human isolates, p<0.001). Similar results were obtained when isolates from hospital patient fecal samples were compared separately with the conventionally raised poultry isolates or when isolates from vegetarian fecal samples were compared separately with isolates from poultry raised without antibiotics (data not shown).
Dendrogram of Extended Virulence Profiles and Phylogenetic Group
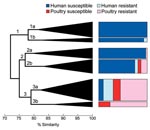
Phylogenetic group and extended virulence profiles among the 243 available ExPEC isolates also were used to construct a similarity dendrogram. The dendrogram showed 3 major clusters, each of which contained 2 prominent subclusters (Figure 3). Isolates were distributed by cluster and subcluster according to source and resistance group; that is, drug-susceptible human isolates accounted for almost all of subclusters 1a, 1b, and 2a. In contrast, drug-resistant human isolates were confined largely to subcluster 3a. Poultry isolates, whether resistant or susceptible, were confined almost entirely to subclusters 2b, 3a, and 3b. Thus, compared with drug-susceptible human isolates, drug-resistant human isolates were significantly more likely to occur within a subcluster, or major cluster, that also contained poultry isolates (p<0.001 for each comparison).
The possible effects of nonindependence among multiple isolates acquired from the same sample were assessed by limiting the analysis to a single isolate per sample, keeping a drug-susceptible isolate (if available) and randomly selecting among multiple drug-resistant isolates where required. This resulted in reduced sample sizes of 681 (total population) and 226 (ExPEC population). The analysis results closely mirrored the pattern of significant findings obtained in the full samples.
In this study, we analyzed the phylogenetic distribution and virulence genotypes of drug-susceptible and drug-resistant E. coli isolates from human volunteers and poultry products in Minnesota and Wisconsin. We found that drug-resistant human isolates, although overlapping somewhat with drug-susceptible human isolates, were more similar overall to poultry isolates than to drug-susceptible human isolates. In contrast, drug-susceptible human isolates differed from poultry isolates. This relationship was observed consistently with diverse analytical approaches and various stratifications of the population. It suggests that many of the drug-resistant human isolates were more likely to have originated in poultry (or a similar nonhuman reservoir) and to have been acquired by humans when these isolates were already drug resistant, than to have emerged de novo in humans by conversion of drug-susceptible human isolates to drug-resistant isolates.
We also found that, regardless of analytical approach and population analyzed, resistant and susceptible poultry isolates were highly similar. This suggests that the resistant poultry isolates likely derived from antimicrobial drug–susceptible, poultry-source E. coli by conversion to resistance. This most plausibly would occur within the avian fecal flora under selection pressure from on-farm use of antimicrobial drugs.
Our findings closely resemble those of a recent study of ciprofloxacin-resistant E. coli from humans and chickens in the late 1990s in Barcelona, Spain (12). These data indicate that these relationships remain valid and are applicable in the United States, to additional resistance phenotypes (specifically quinolones, TMP-SMZ, and extended-spectrum cephalosporins), and to retail poultry products (12). Moreover, similar results were obtained with retail poultry products and poultry carcasses from processing plants. This implies that drug-resistant poultry-source E. coli isolates originate in the birds, rather than being introduced from some exogenous reservoir later during the packaging and distribution process. This in turn suggests that on-farm practices, including use of antimicrobial agents for growth promotion, metaphylaxis, and therapy (21,22), may influence characteristics of E. coli that contaminate retail poultry products and, seemingly, are then transmitted to humans (7).
The greater overall similarity of drug-resistant human isolates to poultry isolates than to drug-susceptible human isolates applied not only to the hospital patient isolates compared with isolates from conventionally raised poultry, but also to the isolates from vegetarians compared with isolates from poultry raised with no antibiotics. This was surprising because the vegetarians ostensibly did not consume poultry and, therefore, should not have been directly exposed to poultry-source E. coli. However, this seeming paradox is consistent with the difficulty in confirming poultry consumption (along with most other individual-level exposures) as an epidemiologic risk factor for colonization with drug-resistant E. coli isolates among community-dwelling persons ([23]; J.R. Johnson, unpub. data). Assuming that the drug-resistant human isolates were derived from poultry, occurrence of poultry-source E. coli in both vegetarians and persons with conventional diets suggests that poultry-source drug-resistant E. coli may spread extensively through the human population without requiring individual exposure to poultry products. This suggestion would be consistent with evidence that household-level risk factors may be more predictive of colonization with drug-resistant E. coli than individual-level risk factors, and that household members often share E. coli clones with each another (23–25). The mechanisms for such diffusion, and methods to block the entry of such strains into the human population and their subsequent spread, need to be defined.
The virulence potential for humans of the present drug-resistant human and poultry E. coli isolates, which is related to their direct threat to human health, is unknown. Predictions regarding virulence potential await molecular comparisons with human clinical isolates (9,10,12) and experimental virulence assessment in vivo (26,27). Nonetheless, the abundance of ExPEC-associated virulence genes in some of these strains is of concern because it suggests a high likelihood of virulence. This would augment any health threat these strains may pose as passive vehicles for drug-resistance genes (6,7).
Potential limitations of this study warrant comment. Because we did not examine alternative sources for drug-resistant human isolates, we cannot exclude the possibility that other foods (28) or nonfood reservoirs (29) might yield even closer similarities to drug-resistant human isolates. Whether persons in the study consumed poultry products from the same lots or suppliers as those sampled is not known. Because the study was conducted in Minnesota and Wisconsin in mostly rural communities and with newly hospitalized patients and nonhospitalized vegetarians, generalizability of the results is unknown. We combined several resistance phenotypes because of low frequencies, which may have obscured differences. We also did not assess other molecular characteristics of strains, e.g., pulsed-field gel electrophoresis profiles (12), sequence types (30), and resistance elements (28). Use of multiple comparisons increased the likelihood of spurious associations (which we addressed by specifying a strict criterion for statistical significance), whereas the small sample size in certain subgroups reduced power for finding true associations.
Strengths of the study include substantial overall sample size, standardized concurrent processing of fecal and poultry samples, close matching of human and poultry samples, extensive molecular typing using virulence-relevant markers, and use of multiple analytical modalities. Additionally, we examined clinically relevant resistance phenotypes.
In summary, our findings suggest that in a contemporary US-based population, many human-source drug-resistant fecal E. coli isolates more likely originated in poultry than in humans, whereas drug-resistant poultry isolates likely derive from drug-susceptible poultry isolates. Our data extend this paradigm to clinically relevant agents other than fluoroquinolones, heighten concerns regarding the potential human health risk for antimicrobial drug use in poultry production, and suggest that avoidance of poultry consumption may not reliably provide personal protection.
Dr Johnson is professor of medicine and director of the Infectious Diseases Fellowship Program at the University of Minnesota and an infectious diseases physician and director of the Molecular Epidemiology Laboratory at the Minneapolis Veterans Affairs Medical Center. His research interests include virulence mechanisms, molecular epidemiology, antimicrobial drug resistance, evolution, reservoirs, and transmission pathways of extraintestinal pathogenic E. coli.
Acknowledgments
We thank Mary Vandermause, Burney Kieke, and Amy Kieke for assistance with the study design, logistics, and data management.
This study was supported by the Office of Research and Development, Medical Research Service, Department of Veterans Affairs; Centers for Disease Control and Prevention grants R01-CI000204 (to J.R.J.) and RS1-CCR520634 (to E.A.B.); the University of Minnesota Rapid Agricultural Response Fund (to J.B.); and the Minnesota Department of Health Emerging Infections program.
References
- Gupta K, Hooton TM, Stamm WE. Increasing antimicrobial resistance and the management of uncomplicated community-acquired urinary tract infections. Ann Intern Med. 2001;135:41–50.PubMedGoogle Scholar
- Pitout JD, Nordmann P, Laupland KB, Poirel L. Emergence of Enterobacteriaciae producing extended spectrum β-lactamases (ESBLs) in the community. J Antimicrob Chemother. 2005;56:52–9. DOIPubMedGoogle Scholar
- Garau J, Xercavins M, Rodriguez-Carballeira M, Gomez-Vera JR, Coll I, Vidal D, Emergence and dissemination of quinolone-resistant Escherichia coli in the community. Antimicrob Agents Chemother. 1999;43:2736–41.PubMedGoogle Scholar
- Russo TA, Johnson JR. Medical and economic impact of extraintestinal infections due to Escherichia coli: an overlooked epidemic. Microbes Infect. 2003;5:449–56. DOIPubMedGoogle Scholar
- Linton AH. Animal to man transmission of Enterobacteriaceae. R Soc Health J. 1977;97:115–8. DOIPubMedGoogle Scholar
- Jones TF, Schaffner W. New perspectives on the persistent scourge of foodborne disease. J Infect Dis. 2005;191:1029–31. DOIPubMedGoogle Scholar
- Collignon P, Angulo FJ. Fluoroquinolone-resistant Escherichia coli: food for thought. J Infect Dis. 2006;194:8–10. DOIPubMedGoogle Scholar
- Johnson JR, Murray AC, Gajewski A, Sullivan M, Snippes P, Kuskowski MA, Isolation and molecular characterization of nalidixic acid-resistant extraintestinal pathogenic Escherichia coli from retail chicken products. Antimicrob Agents Chemother. 2003;47:2161–8. DOIPubMedGoogle Scholar
- Johnson JR, Delavari P, O’Bryan TT, Smith K, Tatini S. Contamination of retail foods, particularly turkey, from community markets (Minnesota, 1999–2000) with antimicrobial-resistant and extraintestinal pathogenic Escherichia coli. Foodborne Pathog Dis. 2005;2:38–49. DOIPubMedGoogle Scholar
- Johnson JR, Kuskowski MA, Smith K, O’Bryan TT, Tatini S. Antimicrobial-resistant and extraintestinal pathogenic Escherichia coli in retail foods. J Infect Dis. 2005;191:1040–9. DOIPubMedGoogle Scholar
- Schroeder CM, White DG, Ge B, Zhang Y, McDermott PF, Avers S, Isolation of antimicrobial-resistant Escherichia coli from retail meats purchased in Greater Washington, DC, USA. Int J Food Microbiol. 2003;85:197–202. DOIPubMedGoogle Scholar
- Johnson JR, Kuskowski MA, Menard M, Gajewski A, Xercavins M, Garau J. Similarity of human and chicken-source Escherichia coli isolates in relation to ciprofloxacin resistance status. J Infect Dis. 2006;194:71–8. DOIPubMedGoogle Scholar
- Mellon M, Benbrook C, Lutz Benbrook K. Hogging it. Estimates of antimicrobial abuse in livestock. Cambridge (MA): UCS Publications; 2001.
- Kieke AL, Borchardt MA, Kieke BA, Spencer SK, Vandermause MF, Smith KE, Use of streptogramin growth promoters in poultry and isolation of streptogramin-resistant Enterococcus faecium from humans. J Infect Dis. 2006;194:1200–8. DOIPubMedGoogle Scholar
- Johnson JR, Murray AC, Kuskowski MA, Schubert S, Prere MF, Picard B, Distribution and characteristics of Escherichia coli clonal group A. Emerg Infect Dis. 2005;11:141–5.PubMedGoogle Scholar
- Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol. 2000;66:4555–8. DOIPubMedGoogle Scholar
- Johnson JR, Stell AL. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J Infect Dis. 2000;181:261–72. DOIPubMedGoogle Scholar
- Gower JC. Some distance properties of latent root and vector methods used in multivariate analysis. Biometrika. 1966;53:325–38.
- Peakall R, Smouse PE. GenAlEx 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes. 2006;6:288–95. DOIGoogle Scholar
- Sokal RR, Sneath PH. Construction of a taxonomic system. In: Sokal RR, Sneath PHA, editors. Principles of numerical taxonomy. San Francisco: W.H. Freeman; 1963.
- Gorbach SL. Antimicrobial use in animal feed--time to stop. N Engl J Med. 2001;345:1202–3. DOIPubMedGoogle Scholar
- Singer RS, Finch R, Wegener HC, Bywater R, Walters J, Lipsitch M. Antibiotic resistance–the interplay between antibiotic use in animals and human beings. Lancet Infect Dis. 2003;3:47–51. DOIPubMedGoogle Scholar
- Hannah EL, Angulo FJ, Johnson JR, Haddadin B, Williamson J, Samore MH. Drug-resistant Escherichia coli, rural Idaho. Emerg Infect Dis. 2005;11:1614–7.PubMedGoogle Scholar
- Murray AC, Kuskowski MA, Johnson JR. Virulence factors predict Escherichia coli colonization patterns among human and animal household members. Ann Intern Med. 2004;140:848–9.PubMedGoogle Scholar
- Manges AR, Johnson JR, Riley LW. Intestinal population dynamics of urinary tract infection-causing Escherichia coli within heterosexual couples. Curr Issues Intest Microbiol. 2004;5:49–57.PubMedGoogle Scholar
- Skyberg JA, Johnson TJ, Johnson JR, Clabots C, Logue CM, Nolan LK. Acquisition of avian pathogenic Escherichia coli plasmids by a commensal E. coli isolate enhances its abilities to kill chicken embryos, grow in human urine, and colonize the murine kidney. Infect Immun. 2006;74:6287–92. DOIPubMedGoogle Scholar
- Johnson JR, Clermont O, Menard M, Kuskowski MA, Picard B, Denamur E. Experimental mouse lethality of Escherichia coli isolates in relation to accessory traits, phylogenetic group, and clinical source. J Infect Dis. 2006;194:1141–50. DOIPubMedGoogle Scholar
- Winokur PL, Vonstein DL, Hoffman EK, Uhlenhopp EK, Doern GV. Evidence for transfer of CMY-2 AmpC β-lactamase plasmids between Escherichia coli and Salmonella isolates from food animals and humans. Antimicrob Agents Chemother. 2001;45:2716–22. DOIPubMedGoogle Scholar
- Johnson JR, Stell A, Delavari P. Canine feces as a reservoir of extraintestinal pathogenic Escherichia coli. Infect Immun. 2001;69:1306–14. DOIPubMedGoogle Scholar
- Tartof SY, Solberg OD, Manges AR, Riley LW. Analysis of a uropathogenic Escherichia coli clonal group by multilocus sequence typing. J Clin Microbiol. 2005;43:5860–4. DOIPubMedGoogle Scholar
Figures
Tables
Cite This ArticleTable of Contents – Volume 13, Number 6—June 2007
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
James R. Johnson, Infectious Diseases (111F), Minneapolis Veterans Affairs Medical Center, 1 Veterans Dr, Minneapolis, MN 55417, USA;
Top