Volume 15, Number 10—October 2009
Dispatch
Escherichia coli as Reservoir for Macrolide Resistance Genes
Abstract
The plasmid-borne mph(A) gene that confers resistance to azithromycin and has recently emerged in Shigella sonnei is present in multidrug- and non–multidrug-resistant Escherichia coli isolates from 4 continents. Further spread of mph(A) to Shigella and Salmonella spp. may be expected.
Macrolides have been regarded for many decades as having good activity and safety for the treatment of infections caused by gram-positive cocci. In general, macrolides show modest potency against Enterobacteriaceae. Most Shigella and Salmonella spp. pathogens display MICs of azithromycin, a macrolide antimicrobial drug, ranging from 2 mg/L to 8 mg/L (1). Despite these relatively high MICs, azithromycin is an attractive option for several reasons. It can be given once a day and attains high intracellular concentrations and sufficient concentrations in the colon of patients to inhibit Shigella and Salmonella spp. Azithromycin is recommended by the American Academy of Pediatrics for treatment of shigellosis in children (2) and by the World Health Organization as a second-line treatment for adults (3). It has also been proposed for short-course treatment of typhoid fever (4).
We recently reported an outbreak of shigellosis in Paris, France; failure of azithromycin treatment was related to emergence of plasmid-mediated resistance to macrolides (5). Resistance was related to the expression of a macrolide 2′ phosphotransferase encoded by the mph(A) gene. Because shigellosis remains a common gastrointestinal disease in both developing and industrialized countries, emergence of macrolide resistance may have major public health consequences.
Since the early reports by Ochiai (6) and Akiba (7) at the end of the 1950s, plasmid-mediated transfer of resistance genes between Escherichia coli and Shigella spp. has been reported in several instances (8). Therefore, we hypothesized that E. coli might constitute a major reservoir for macrolide resistance genes that could be subsequently transferred to Shigella sonnei.
Acquired resistance to macrolides may result from a variety of mechanisms of resistance, several of which have already been reported in Enterobacteriaceae (9,10). These mechanisms include target site modification by methylases encoded by erm genes, in particular erm(A), erm(B), and erm(C). Macrolides may be inactivated by modifying enzymes first reported in Enterobacteriaceae (11,12), e.g., esterases encoded by ere(A) or ere(B) genes or phosphotransferases encoded by mph(A), mph(B), and mph(D) genes. The third mechanism is acquisition of efflux pumps, mef(A) and msr(A), that have been found essentially in gram-positive organisms, although mef(A) has been identified in gram-negative organisms (10). All of these genes confer full cross-resistance between erythromycin and azithromycin (9). We aimed to assess the prevalence of acquired resistance to macrolides in commensal and clinical isolates of E. coli from various geographic origins and to characterize the mechanisms underlying E. coli resistance to macrolides.
A total of 190 E. coli isolates were collected from 5 countries on 4 continents. Some of these isolates were obtained from populations exposed to low antimicrobial selective pressure; 45 commensal isolates were from feces of healthy Wayampi Amerindians in French Guiana, 20 from feces of children living in a remote village of Senegal, and 49 from feces of healthy nurses working in a hospital in Paris. Other isolates were obtained from populations having received multiple antimicrobial drug treatments; 29 isolates were from feces of children from Niger hosted in a center for nutritional rehabilitation, and 47 isolates were producers of extended-spectrum β-lactamase (ESBL) obtained from various clinical samples in hospitalized patients in Vietnam (n = 37) and France (Hospital of Caen) (n = 10).
Susceptibility to 16 antimicrobial drugs was determined by the disk-diffusion method. MICs of erythromycin were determined by the agar dilution technique, and ESBLs were detected by the double-disk synergy test, as recommended by the French Society for Microbiology (www.sfm.asso.fr).
E. coli isolates from French Guiana, Senegal, and Paris were susceptible to quinolones, gentamicin, and third-generation cephalosporins. Resistance to amoxicillin-ticarcillin (by penicillinase production) was detected for 22.2%, 20.4%, and 40.0% of the isolates obtained from nurses in Guiana, Paris, and Senegal, respectively. Coresistance to amoxicillin and cotrimoxazole was found for 13%, 14%, and 35% of isolates, respectively.
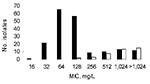
Multidrug-resistant isolates were commonly obtained from Niger natives; 34.4% were resistant to both cefotaxime (mostly by ESBL production) and ciprofloxacin, and 58.6% to gentamicin. ESBL producers from Vietnam and Caen hospital displayed resistance to ciprofloxacin for 86.5% and 60.0% and resistance to gentamicin for 86.4% and 50.0% of isolates, respectively. MICs of erythromycin ranged from 16 mg/L to >1,024 mg/L (Table 1; Figure). Distribution of MICs of erythromycin was bimodal; mode = 64 mg/L for 1 population with low MICs and >1,024 mg/L for the other population with high MICs (Figure).
MICs differed according to the origin of the isolates. Multiple resistance was associated with MICs of erythromycin >256 mg/L with 1 exception: an isolate from Guiana was resistant only to amoxicillin and cotrimoxazole (MIC of erythromycin, 1,024 mg/L).
We screened for macrolide resistance genes by using oligonucleotide primers and PCR conditions (Table 2). PCR reactions were performed as follows: an initial denaturation step (95oC, 3 min) followed by 30 cycles consisting of denaturation (95oC, 30 s), annealing at a temperature depending on the primers used (30 s), elongation (72oC, 30 s) and a final extension step (72oC, 10 min). Positive and negative controls were included in each run.
The mph(A) gene was commonly present in 34 isolates (MICs 256 mg/L to >1,024 mg/L). The gene was mostly detected in isolates resistant to cefotaxime (27 isolates) but also in 4 (21%) of 19 isolates resistant to only amoxicillin and cotrimoxazole in different countries. To confirm this latter association, we searched for the mph(A) gene in 100 clinical isolates of E. coli from the Caen hospital coresistant to amoxicillin and cotrimoxazole but susceptible to cefotaxime, which is a common phenotype present in ≈25% of E. coli isolates from this hospital. The gene was detected in 13 isolates (MIC >256 mg/L), confirming the presence of the gene in non–multidrug-resistant E. coli (Table 1). In a previous study on the distribution of 7 macrolide resistance genes in gram-negative isolates from the urine and oral cavity of healthy children in Portugal, Ojo et al. detected the mph(A) gene in 15 of 26 studied E. coli isolates (10). However, the profile of resistance to other antimicrobial drugs was not determined.
The other macrolide resistance genes were more scarce. The erm(B) gene was detected in 2 isolates (MICs >1,024 mg/L) and mph(B) in 2 others (MICs 128 mg/L). In 4 isolates (MICs >1,024 mg/L), both mph(A) and erm(B) were amplified. The 6 other genes, erm(A), erm(C), ere(A), ere(B), mef(A), and msr(A), were not detected. In 6 isolates with MICs of erythromycin equal to 256 mg/L and 2 with MICs of erythromycin equal to 512 mg/L, no resistance gene could be amplified, suggesting the presence of other macrolide resistance determinants. Distribution of the resistance genes mph(A), erm(B), and mph(B) is shown in Table 1 and in the Figure.
The plasmid-borne mph(A) gene detected in S. sonnei resistant to azithromycin was the most common macrolide resistance gene detected in E. coli collected in 5 countries on 4 continents. The gene was mostly detected in isolates from patients who had received antimicrobial drugs or had been hospitalized, in particular in ESBL producers, but was also detected in E. coli isolates coresistant to amoxicillin and cotrimoxazole, which are common worldwide. Because E. coli and Shigella spp. are phylogenetically closely related species that easily exchange plasmids, further dissemination of resistance to macrolides in the latter species may be predicted. Also, plasmid-mediated resistance to macrolides may emerge in Salmonella spp., which is also a target of azithromycin.
Ms Nguyen is pursuing a master’s degree at the University of Caen. The focus of her work is the identification of reservoirs of macrolide resistance genes.
References
- Gordillo ME, Singh KV, Murray B. In vitro activity of azithromycin against bacterial enteric pathogens. Antimicrob Agents Chemother. 1993;37:1203–5.PubMedGoogle Scholar
- Centers for Disease Control and Prevention. Outbreaks of multidrug-resistant Shigella sonnei gastroenteritis associated with day care centers—Kansas, Kentucky, and Missouri, 2005. MMWR Morb Mortal Wkly Rep. 2006;55:1068–71.PubMedGoogle Scholar
- World Health Organization. Guidelines for the control of shigellosis including epidemics due to Shigella dysenteriae type 1 [cited 23 Mar 2009]. Available from http://www.who.int/child-adolescent-health/publications/pubemergencies.htm
- Effa EE, Bukirwa H. Azithromycin for treating uncomplicated typhoid and paratyphoid fever (enteric fever). Cochrane Database Syst Rev. 2008;4:CD006083.PubMedGoogle Scholar
- Boumghar-Bourtchai L, Mariani-Kurkdjian P, Bingen E, Filliol I, Dhalluin A, Ifrane SA, Macrolide-resistant Shigella sonnei. Emerg Infect Dis. 2008;14:1297–9. DOIPubMedGoogle Scholar
- Ochiai K, Yamanaka T, Kimura K, Sawada 0. Studies on transfer of drug resistance between Shigella strains and Escherichia coli strains [in Japanese]. Nihon-ijishimpo No. 1959;1861:34–46.
- Akiba T, Koyama T, Isshiki T, Kimura S, Fukushima T. On the mechanism of the development of multiple-drug-resistant clones of Shigella. Jpn J Microbiol. 1960;4:219–27.PubMedGoogle Scholar
- Tauxe RV, Cavanagh TR, Cohen ML. Interspecies gene transfer in vivo producing an outbreak of multiple resistant shigellosis. J Infect Dis. 1989;160:1067–70.PubMedGoogle Scholar
- Leclercq R. Mechanisms of resistance to macrolides and lincosamides: nature of the resistance elements and their clinical implications. Clin Infect Dis. 2002;34:482–92. DOIPubMedGoogle Scholar
- Ojo KK, Ulep C, Van Kirk N, Luis H, Bernardo M, Leitao J, The mef(A) gene predominates among seven macrolide resistance genes identified in gram-negative strains representing 13 genera, isolated from healthy Portuguese children. Antimicrob Agents Chemother. 2004;48:3451–6. DOIPubMedGoogle Scholar
- Arthur M, Andremont A, Courvalin P. Distribution of erythromycin esterase and rRNA methylase genes in members of the family Enterobacteriaceae highly resistant to erythromycin. Antimicrob Agents Chemother. 1987;31:404–9.PubMedGoogle Scholar
- O'Hara K, Kanda T, Ohmiya K, Ebisu T, Kono M. Purification and characterization of macrolide 2'-phosphotransferase from a strain of Escherichia coli that is highly resistant to erythromycin. Antimicrob Agents Chemother. 1989;33:1354–7.PubMedGoogle Scholar
Figure
Tables
Cite This ArticleTable of Contents – Volume 15, Number 10—October 2009
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Roland Leclercq, CHU de Caen, Service de Microbiologie, ave Côte de Nacre, 14033 Caen CEDEX, France
Top