New Respiratory Enterovirus and Recombinant Rhinoviruses among Circulating Picornaviruses
Caroline Tapparel
1
, Thomas Junier
1, Daniel Gerlach, Sandra Van Belle, Lara Turin, Samuel Cordey, Kathrin Mühlemann, Nicolas Regamey, John-David Aubert, Paola M. Soccal, Philippe Eigenmann, Evgeny M. Zdobnov
1, and Laurent Kaiser
1
Author affiliations: University of Geneva Hospitals, Geneva, Switzerland (C. Tapparel, S. Van Belle, L. Turin, S. Cordey, P. M. Soccal, P. Eigenmann, L. Kaiser); University of Geneva Medical School, Geneva (C. Tapparel, T. Junier, D. Gerlach, S. Van Belle, L. Turin, S. Cordey, E. Zdobnov, L. Kaiser); Swiss Institute of Bioinformatics, Geneva (T. Junier, D. Gerlach, E. Zdobnov); University Hospital of Bern, Bern, Switzerland (K. Mühlemann, N. Regamey); University Hospital of Lausanne, Lausanne, Switzerland (J.-D. Aubert); Imperial College London, London, UK (E. Zdobnov); 1These authors contributed equally to this article.
Main Article
Figure 3
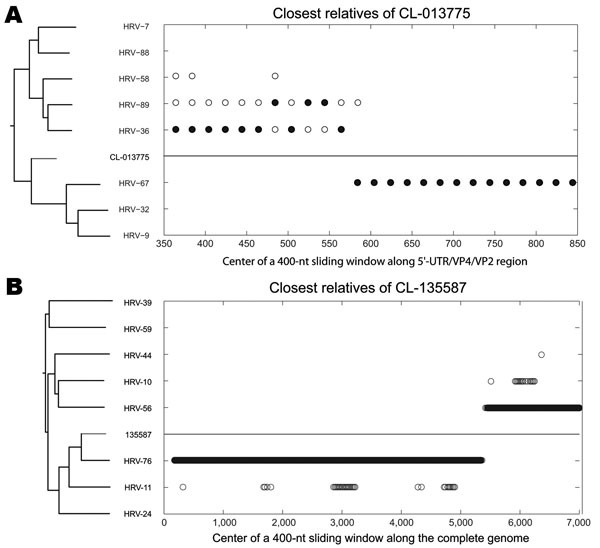
Figure 3. Nearest-neighbor relatedness of rhinovirus CL-013775 (and CL-073908) along the 5′ untranslated region/VP4/VP2 region (A), and nearest-neighbor relatedness of rhinovirus CL-135587 along the complete genome (B), identified by bootscanning. At each position of a sliding window, the solid circles indicate the closest relative within a defined threshold of the phylogenetic distance to CL-013775 (A) and CL-135587 (B). Both panels show phylogenetic trees of analyzed serotypes over the entire scanned region. Human rhinovirus 7 (HRV-7), -9, -10, -11, -24, -32 (accession nos. EU096019, AF343584), -36, -39, -44, -56, -58 (EU096045, AY040236), -59, -67 (EU096054, AF343603, and DQ473505), -76, -88, and -89 sequences were obtained from GenBank (see Technical Appendix 2 Figure 1, for full-length genome accession numbers).
Main Article
Page created: December 16, 2010
Page updated: December 16, 2010
Page reviewed: December 16, 2010
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
