Volume 15, Number 9—September 2009
Research
Genetics and Pathogenesis of Feline Infectious Peritonitis Virus
Figure 5
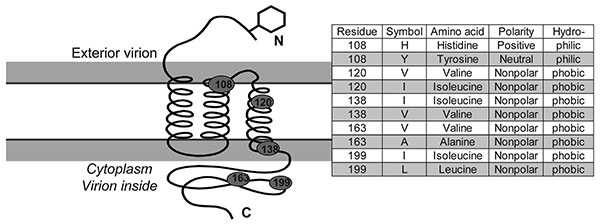
Figure 5. Diagram of membrane protein containing 3 transmembrane helices, an external N terminus and an internal carboxy terminus. Approximate position of 5 variable diagnostic amino acid sites (see Table 2) as determined by sequence comparison to severe acute respiratory syndrome coronavirus (32). Amino acid residue, polarity, and hydrophobicity or hydropholicity is stated.
References
- Addie DD. Clustering of feline coronaviruses in multicat households. Vet J. 2000;159:8–9. DOIPubMedGoogle Scholar
- Addie DD, Jarrett O. A study of naturally occurring feline coronavirus infections in kittens. Vet Rec. 1992;130:133–7.PubMedGoogle Scholar
- Kennedy M, Citino S, McNabb AH, Moffatt AS, Gertz K, Kania S. Detection of feline coronavirus in captive Felidae in the USA. J Vet Diagn Invest. 2002;14:520–2.PubMedGoogle Scholar
- Pedersen NC. A review of feline infectious peritonitis virus infection: 1963–2008. J Feline Med Surg. 2009;11:225–58. DOIPubMedGoogle Scholar
- Foley JE, Poland A, Carlson J, Pedersen NC. Risk factors for feline infectious peritonitis among cats in multiple-cat environments with endemic feline enteric coronavirus. J Am Vet Med Assoc. 1997;210:1313–8.PubMedGoogle Scholar
- Pedersen NC, Evermann JF, McKeirnan AJ, Ott RL. Pathogenicity studies of feline coronavirus isolates 79-1146 and 79-1683. Am J Vet Res. 1984;45:2580–5.PubMedGoogle Scholar
- de Groot RJ. Feline infectous peritonitis. In: Siddell SG, editor. The Coronoviridae. New York: Plenum Press; 1995. p. 293–309.
- Weiss RC, Scott FW. Pathogenesis of feline infectious peritonitis: nature and development of viremia. Am J Vet Res. 1981;42:382–90.PubMedGoogle Scholar
- Kipar A, Kohler K, Leukert W, Reinacher M. A comparison of lymphatic tissues from cats with spontaneous feline infectious peritonitis (FIP), cats with FIP virus infection but no FIP, and cats with no infection. J Comp Pathol. 2001;125:182–91. DOIPubMedGoogle Scholar
- Kipar A, Meli ML, Failing K, Euler T, Gomes-Keller MA, Schwartz D, Natural feline coronavirus infection: differences in cytokine patterns in association with the outcome of infection. Vet Immunol Immunopathol. 2006;112:141–55. DOIPubMedGoogle Scholar
- Hunziker L, Recher M, Macpherson AJ, Ciurea A, Freigang S, Hengartner H, Hypergammaglobulinemia and autoantibody induction mechanisms in viral infections. Nat Immunol. 2003;4:343–9. DOIPubMedGoogle Scholar
- Poland AM, Vennema H, Foley JE, Pedersen NC. Two related strains of feline infectious peritonitis virus isolated from immunocompromised cats infected with a feline enteric coronavirus. J Clin Microbiol. 1996;34:3180–4.PubMedGoogle Scholar
- Vennema H, Poland A, Foley J, Pedersen NC. Feline infectious peritonitis viruses arise by mutation from endemic feline enteric coronaviruses. Virology. 1998;243:150–7. DOIPubMedGoogle Scholar
- Rottier PJ, Nakamura K, Schellen P, Volders H, Haijema BJ. Acquisition of macrophage tropism during the pathogenesis of feline infectious peritonitis is determined by mutations in the feline coronavirus spike protein. J Virol. 2005;79:14122–30. DOIPubMedGoogle Scholar
- Stoddart CA, Scott FW. Intrinsic resistance of feline peritoneal macrophages to coronavirus infection correlates with in vivo virulence. J Virol. 1989;63:436–40.PubMedGoogle Scholar
- Haijema BJ, Volders H, Rottier PJ. Switching species tropism: an effective way to manipulate the feline coronavirus genome. J Virol. 2003;77:4528–38. DOIPubMedGoogle Scholar
- Can-Sahna K, Soydal Ataseven V, Pinar D, Oguzoglu TC. The detection of feline coronaviruses in blood samples from cats by mRNA RT-PCR. J Feline Med Surg. 2007;9:369–72. DOIPubMedGoogle Scholar
- Dye C, Siddell SG. Genomic RNA sequence of feline coronavirus strain FCoV C1Je. J Feline Med Surg. 2007;9:202–13. DOIPubMedGoogle Scholar
- Hartley O, Klasse PJ, Sattentau QJ, Moore JP. V3: HIV's switch-hitter. AIDS Res Hum Retroviruses. 2005;21:171–89. DOIPubMedGoogle Scholar
- Ballesteros ML, Sanchez CM, Enjuanes L. Two amino acid changes at the N-terminus of transmissible gastroenteritis coronavirus spike protein result in the loss of enteric tropism. Virology. 1997;227:378–88. DOIPubMedGoogle Scholar
- Saif LJaS. K. Transmissible gastroenteritis virus and porcine respiratory coronavirus. In: Zimmerman JJ, editor. Diseases of swine. 9th ed. Ames (IA): Iowa State University Press; 2006. p. 489–516.
- Sanchez CM, Izeta A, Sanchez-Morgado JM, Alonso S, Sola I, Balasch M, Targeted recombination demonstrates that the spike gene of transmissible gastroenteritis coronavirus is a determinant of its enteric tropism and virulence. J Virol. 1999;73:7607–18.PubMedGoogle Scholar
- Mongkolsapaya J, Dejnirattisai W, Xu XN, Vasanawathana S, Tangthawornchaikul N, Chairunsri A, Original antigenic sin and apoptosis in the pathogenesis of dengue hemorrhagic fever. Nat Med. 2003;9:921–7. DOIPubMedGoogle Scholar
- Anishchenko M, Bowen RA, Paessler S, Austgen L, Greene IP, Weaver SC. Venezuelan encephalitis emergence mediated by a phylogenetically predicted viral mutation. Proc Natl Acad Sci U S A. 2006;103:4994–9. DOIPubMedGoogle Scholar
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–82. DOIPubMedGoogle Scholar
- Maddison DRaM. W.P. MacClade 3.05. Sunderland (MA): Sinauer; 1995.
- Swofford DL. PAUP*: Phylogenetic Analysis Using Parsimony (*and other methods). Sunderland (MA): Sinauer; 2002.
- Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–8. DOIPubMedGoogle Scholar
- Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004;5:150–63. DOIPubMedGoogle Scholar
- Pearks Wilkerson AJ, Teeling EC, Troyer JL, Bar-Gal GK, Roelke M, Marker L, Coronavirus outbreak in cheetahs: lessons for SARS. Curr Biol. 2004;14:R227–8. DOIPubMedGoogle Scholar
- Rottier PJ. The coronavirus membrane glycoprotein. In: Siddell SG, editor. The Coronaviridae. New York: Plenum Press; 1995. p. 115–40.
- He Y, Zhou Y, Siddiqui P, Niu J, Jiang S. Identification of immunodominant epitopes on the membrane protein of the severe acute respiratory syndrome-associated coronavirus. J Clin Microbiol. 2005;43:3718–26. DOIPubMedGoogle Scholar
- Heeney JL, Evermann JF, McKeirnan AJ, Marker-Kraus L, Roelke ME, Bush M, Prevalence and implications of feline coronavirus infections of captive and free-ranging cheetahs (Acinonyx jubatus). J Virol. 1990;64:1964–72.PubMedGoogle Scholar
- Pontius JU, Mullikin JC, Smith DR, Lindblad-Toh K, Gnerre S, Clamp M, Initial sequence and comparative analysis of the cat genome. Genome Res. 2007;17:1675–89. DOIPubMedGoogle Scholar
Page created: December 07, 2010
Page updated: December 07, 2010
Page reviewed: December 07, 2010
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.