Volume 16, Number 4—April 2010
Research
Livestock-associated Methicillin-Resistant Staphylococcus aureus Sequence Type 398 in Humans, Canada
Figure 2
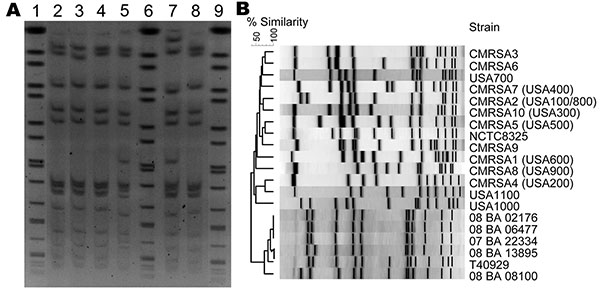
Figure 2. A) Pulsed-field gel electrophoresis (PFGE) of Cfr91-digested livestock-associated methicillin-resistant Staphylococcus aureus (MRSA). Lanes 1, 6, and 9, universal standard Salmonella Braenderup H9812; Lane 2, 08 BA 02176; Lane 3, 08 BA 13895; Lane 4, 07 BA 06477; Lane 5, T40929; Lane 7, 08 BA 08100; Lane 8, 07 BA 22334. B) PFGE dendrogram comparing the Cfr91 fingerprint patterns of 6 livestock-associated MRSA isolates from humans in Canada with the SmaI fingerprints of other human epidemic strains of MRSA circulating in Canada.
Page created: December 28, 2010
Page updated: December 28, 2010
Page reviewed: December 28, 2010
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.