Volume 17, Number 10—October 2011
Dispatch
Novel Arenavirus, Zambia
Figure 1
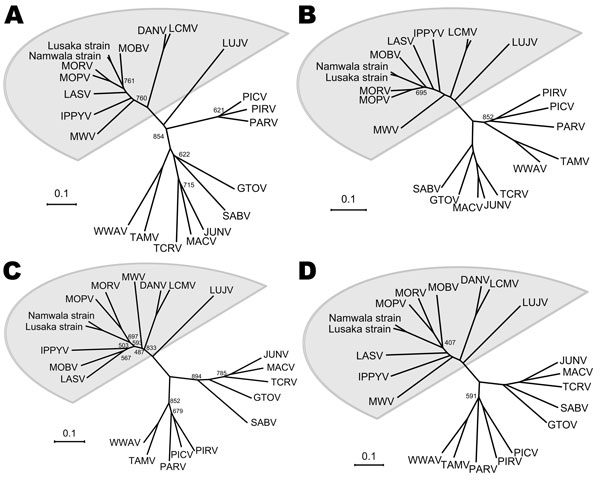
Figure 1. Phylogenetic analysis of Luna virus proteins based on the amino acid sequence, Zambia, 2009. Phylogenetic tree of A) glycoprotein precursor (GPC), B) nucleoprotein (NP), C) Z protein, and D) L protein. Bootstrap values are indicated in the trees (<900). Data from which amino acid sequences used for phylogenetic analyses were deduced: DANV, Dandenong virus (GenBank accession nos. EU136038 and EU136039); IPPYV, Ippy virus (GenBank accession nos. NC_007905 and NC_007906); LASV, Lassa virus (GenBank accession nos. NC_004296 and NC_004297); LUJV, Lujo virus (GenBank accession nos. FJ952384 and FJ952385); LCMV, lymphocytic choriomeningitis virus (GenBank accession nos. EU480450 and EU480453); MWV, Merino Walk virus (GenBank accession nos. GU078660 and GU078661); MOBV, Mobala virus (GenBank accession nos. NC_007903 and NC_007904); MOPV, Mopeia virus (GenBank accession nos. NC_006575 and NC_006574); MORV, Morogoro virus (GenBank accession nos. NC_013057 and NC_013058); GTOV, Guanarito virus (GenBank accession nos. AY129247 and AY358024); JUNV, Junin virus (GenBank accession nos. AY746353 and AY746354); MACV, Machupo virus (GenBank accession nos. AY624355 and AY624354); PARV, Parana virus (GenBank accession nos. AF512829 and NC_010761); PICV, Pichinde virus (GenBank accession nos.EF529746 and EF529747); PIRV, Pirital virus (GenBank accession nos. EU542420 and NC_005897); SABV, Sabia virus (GenBank accession nos. NC_006317 and NC_006313); TCRV, Tacaribe virus (GenBank accession nos. NC_004293 and NC_004292); TAMV, Tamiami virus (GenBank accession nos.NC_010701 and NC_010702); and WWAV, Whitewater Arroyo virus (GenBank accession nos. NC_010700 and [NC_0107003). Scale bars indicate amino acid substitutions per site.