Volume 17, Number 3—March 2011
Dispatch
Multidrug-Resistant Genotypes of Plasmodium falciparum, Myanmar
Cite This Article
Citation for Media
Abstract
We performed a molecular epidemiologic survey of mutations associated with drug-resistance genes in Plasmodium falciparum in northeastern Myanmar. In this region, 3 highly mutated drug-resistance haplotypes and 1 associated with decreased quinine susceptibility were prevalent, which suggests that parasites may be resistant to multiple commonly used antimalarial drugs.
Malaria is a major impediment to socioeconomic development in the Greater Mekong Subregion (GMS) of Southeast Asia (1). Malaria distribution in the GMS is extremely uneven, with areas of high endemicity in some countries and along international borders. In Myanmar, malaria is particularly problematic; more than half of malaria cases and approximately three fourths of malaria-related deaths in the GMS during 2007 occurred in Myanmar. The GMS has been the breeding ground of multidrug-resistant Plasmodium falciparum, and resistance to chloroquine and antifolates arose there and spread to Africa (2,3). In particular, recent detection of reduced artemisinin susceptibility at the Thailand–Cambodia border is a major concern (4). As a result, drug resistance has been monitored extensively in this region. In contrast, information about resistance to antimalarial drugs in Myanmar is exceptionally scarce. Accordingly, as our initial step toward a comprehensive antimalarial drug study in Myanmar, we performed a molecular survey of drug resistance in the northeastern region of this country.
We screened by microscopy 4,980 patients with febrile illness who sought care at a malaria clinic in Kachin State, northeastern Myanmar, during 2007–2009; a total of 27.9% had malaria infections. P. falciparum, P. vivax, and mixed species infections accounted for 56.7%, 41.1%, and 2.2% of malaria cases, respectively. Finger-prick blood samples were obtained from 260 patients with uncomplicated P. falciparum infection who had not used antimalarial drugs during the previous 2 weeks. Parasite DNA was extracted from filter papers and genotyped at 3 polymorphic genes, which detected 54.6% of samples containing mixed-strain infections (5). Of the 118 samples with monoclonal infection, 117 samples were successfully genotyped by PCR and sequencing at 5 known and putative drug-resistance genes (Technical Appendix), some of which have been widely used for resistance surveillance and as predictors of clinical efficacy of antimalarial drugs.
Sequencing of 2 fragments in the P. falciparum chloroquine resistance transporter (pfcrt) gene covering single nucleotide polymorphisms (SNPs) at codons 72–76 and 220, respectively (6), showed that the major chloroquine resistance determinant K76T mutation has reached fixation in the parasite population (Figure 1). All parasites had sequence CVIET at positions 72–76, compared with the wild-type sequence SVMNK. In addition, the A220S mutation associated with chloroquine resistance was predominant (99.1%).
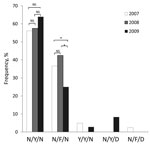
Sequencing of 2 P. falciparum multidrug resistance 1 (pfmdr1) fragments as described (5) detected only the N86Y, Y184F, and N1042D mutations making up 5 haplotypes (Figure 2). The overall haplotype prevalence differed significantly among the years (p<0.001, χ2 = 31.39, df = 8).The prevalence of wild-type haplotype was 59.0%. Among the 3 mutant codons, only Y184F reached a relatively high frequency (35.9%). One sample contained the double mutations 184F/1042D. Analysis of pfmdr1 copy number from monoclonal infections by real-time PCR with 3D7 and Dd2 strains as negative and positive controls, respectively (6), detected no pfmdr1 amplification.
Sequencing of the dihydrofolate reductase (pfdhfr) and dihydropoteroate synthase (pfdhps) genes (Technical Appendix) detected 4 SNPs in pfdhfr associated with pyrimethamine resistance (N51I, C59R, S108N, and I164L) and 4 SNPs in pfdhps associated with sulfadoxine resistance (S436A, A437G, K540E/N, and A581G) (Table 1). The overall haplotype prevalence of the 2 genes differed significantly between the years (p<0.0001, χ2 = 76.49, df = 28). Of the 5 pfdhfr haplotypes, wild-type NCSI was observed only in 1 sample in 2007; the remaining samples contained at least double mutations 59R/108N. Two triple-mutation haplotypes (NRNL and IRNI, mutations in boldface) were detected with NRNL being more frequent than IRNI in each year. Overall, quadruple mutations (IRNL) were found in >50% of the samples. In addition, frequency of triple and quadruple mutations increased gradually from 2007 to 2009. We found all 5 haplotypes in 2007 but only triple and quadruple mutations in 2009. In pfdhps, 10 haplotypes were found, and 437G and 540E/N mutations were highly prevalent: 98.3 and 96.6%, respectively (Table 1). Similarly, the wild-type pfdhps haplotype SGKA was found in only 2 samples. AGEA was the most common haplotype in each year and reached an overall frequency of 48.7%. Quadruple mutations (AGEG) were found only in 2008 and 2009.
Molecular analysis of drug-resistance markers in monoclonal infections enabled us to obtain multilocus genotypes of the parasites. Genotyping each of the 117 parasite isolates at 16 drug resistance–related codons in the pfcrt, pfmdr1, pfdhfr, and pfdhps genes showed 41 haplotypes (Figure 1). Among these haplotypes, parasites containing >10 mutated codons accounted for 93.2% of the samples.
Polymorphisms in the minisatellite ms4760 of P. falciparum Na+/H+ exchanger (pfnhe1) are associated with quinine sensitivity (5,7–10). Sequencing of the pfnhe1 fragment containing the ms4760 minisatellite from 79 monoclonal infections showed 10 haplotypes, with haplotype 7 the most predominant (54.4%) (Table 2). More than 64% of samples tested contained >3 copies of the DNNND repeat (Rep1); 76% contained 1 copy of the NHNDNHNNDDD repeat (Rep2). Accordingly, >60% of parasite isolates had a Rep1:Rep2 ratio of >3:1.
In Myanmar, high-level resistance to chloroquine and pyrimethamine–sulfadoxine was reported more than a decade ago (11–13). Our molecular survey showed that the major chloroquine resistance allele CVIET has reached fixation, and triple and quadruple mutations in pfdhfr and pfdhps were highly prevalent in this region. These findings strongly suggest that a large proportion of parasites might show clinical resistance to chloroquine and antifolate drugs. Although chloroquine has been withdrawn from treating P. falciparum malaria for decades in some regions, the pfcrt resistance alleles showed no sign of abating (6). Furthermore, despite adoption of artemisinin combination therapy in 2002, the frequency of highly mutated pfdhfr and pfdhps haplotypes appeared to have increased during this study, which suggested that artemisinin combination therapy might not have retarded the spread of antifolate-resistant parasites. This situation differs from that in the western Myanmar border area but is similar to that in Thailand and Cambodia, where highly mutated pfdhfr and pfdhps genotypes also were common (14,15).
Mutations in pfmdr1 are associated with resistance to several antimalarial drugs including chloroquine, mefloquine, and quinine and increased pfmdr1 copy number is responsible for mefloquine resistance. We found that ≈60% of the parasites contained the wild-type pfmdr1 allele, similar to some parasites from the western Myanmar border area (15). No pfmdr1 amplification was detected, suggesting that parasites from this region might be mefloquine sensitive, consistent with the fact that mefloquine has not been deployed here. In contrast, in vitro mefloquine resistance was observed in southeast Myanmar bordering Thailand (13), possibly because of the extensive use of mefloquine in Thailand for the past 2 decades.
Although the validity of pfnhe1 minisatellite polymorphism for predicting quinine resistance remains uncertain and may depend on the parasites’ origins (7–9), we detected significant association of decreased quinine susceptibility with increased DNNND repeat copies (5). We have provided further evidence on the high prevalence of parasites with increased DNNND repeats in pfnhe1, which suggests that some parasite strains might show reduced sensitivity to quinine.
Overall, our molecular survey of antimalarial drug resistance in P. falciparum showed high frequency of multidrug-resistant haplotypes in northeastern Myanmar. Moreover, parasites in this region had unique multilocus genotypes that differed markedly from those in other areas of the GMS. These findings suggest that coordinated efforts are necessary to thwart the spread of resistant strains across larger geographic regions. Our molecular study showed only the genotypes of the drug resistance genes; further in vitro and in vivo studies are required to corroborate these findings.
Dr Yang is a professor at the Department of Parasitology, Kunming Medical University, Yunnan, People’s Republic of China. Her primary research interests are epidemiology of drug resistance in malaria parasites.
Acknowledgments
We thank Guofa Zhou for assistance with statistical analysis.
This study was supported by 1R01AI075429 and 1U19AI089672 from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, and by grant no. 30960050 from the National Natural Science Foundation of China.
References
- Socheat D, Denis MB, Fandeur T, Zhang Z, Yang H, Xu J, Mekong malaria. II. Update of malaria, multi-drug resistance and economic development in the Mekong region of Southeast Asia. Southeast Asian J Trop Med Public Health. 2003;34(Suppl 4):1–102.PubMedGoogle Scholar
- Wootton JC, Feng X, Ferdig MT, Cooper RA, Mu J, Baruch DI, Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum. Nature. 2002;418:320–3. DOIPubMedGoogle Scholar
- Roper C, Pearce R, Nair S, Sharp B, Nosten F, Anderson T. Intercontinental spread of pyrimethamine-resistant malaria. Science. 2004;305:1124. DOIPubMedGoogle Scholar
- Noedl H, Se Y, Schaecher K, Smith BL, Socheat D, Fukuda MM. Evidence of artemisinin-resistant malaria in western Cambodia. N Engl J Med. 2008;359:2619–20. DOIPubMedGoogle Scholar
- Meng H, Zhang R, Yang H, Fan Q, Su X, Miao J, In vitro sensitivity of Plasmodium falciparum clinical isolates from the China–Myanmar border area to quinine and association with polymorphism in the Na+/H+ exchanger. Antimicrob Agents Chemother. 2010;54:4306–13. DOIPubMedGoogle Scholar
- Yang Z, Zhang Z, Sun X, Wan W, Cui L, Zhang X, Molecular analysis of chloroquine resistance in Plasmodium falciparum in Yunnan Province, China. Trop Med Int Health. 2007;12:1051–60. DOIPubMedGoogle Scholar
- Henry M, Briolant S, Zettor A, Pelleau S, Baragatti M, Baret E, Plasmodium falciparum Na+/H+ exchanger 1 transporter is involved in reduced susceptibility to quinine. Antimicrob Agents Chemother. 2009;53:1926–30. DOIPubMedGoogle Scholar
- Okombo J, Kiara SM, Rono J, Mwai L, Pole L, Ohuma E, In vitro activities of quinine and other antimalarials and pfnhe polymorphisms in Plasmodium isolates from Kenya. Antimicrob Agents Chemother. 2010;54:3302–7. DOIPubMedGoogle Scholar
- Andriantsoanirina V, Menard D, Rabearimanana S, Hubert V, Bouchier C, Tichit M, Association of microsatellite variations of Plasmodium falciparum Na+/H+ exchanger (Pfnhe-1) gene with reduced in vitro susceptibility to quinine: lack of confirmation in clinical isolates from Africa. Am J Trop Med Hyg. 2010;82:782–7. DOIPubMedGoogle Scholar
- Ferdig MT, Cooper RA, Mu J, Deng B, Joy DA, Su XZ, Dissecting the loci of low-level quinine resistance in malaria parasites. Mol Microbiol. 2004;52:985–97. DOIPubMedGoogle Scholar
- Smithuis FM, Monti F, Grundl M, Oo AZ, Kyaw TT, Phe O, Plasmodium falciparum: sensitivity in vivo to chloroquine, pyrimethamine/sulfadoxine and mefloquine in western Myanmar. Trans R Soc Trop Med Hyg. 1997;91:468–72. DOIPubMedGoogle Scholar
- Smithuis F, Shahmanesh M, Kyaw MK, Savran O, Lwin S, White NJ. Comparison of chloroquine, sulfadoxine/pyrimethamine, mefloquine and mefloquine-artesunate for the treatment of falciparum malaria in Kachin State, north Myanmar. Trop Med Int Health. 2004;9:1184–90. DOIPubMedGoogle Scholar
- Wongsrichanalai C, Lin K, Pang LW, Faiz MA, Noedl H, Wimonwattrawatee T, In vitro susceptibility of Plasmodium falciparum isolates from Myanmar to antimalarial drugs. Am J Trop Med Hyg. 2001;65:450–5.PubMedGoogle Scholar
- Khim N, Bouchier C, Ekala MT, Incardona S, Lim P, Legrand E, Countrywide survey shows very high prevalence of Plasmodium falciparum multilocus resistance genotypes in Cambodia. Antimicrob Agents Chemother. 2005;49:3147–52. DOIPubMedGoogle Scholar
- Anderson TJ, Nair S, Sudimack D, Williams JT, Mayxay M, Newton PN, Geographical distribution of selected and putatively neutral SNPs in Southeast Asian malaria parasites. Mol Biol Evol. 2005;22:2362–74. DOIPubMedGoogle Scholar
Figures
Tables
Cite This Article1These authors contributed equally to this article.
Table of Contents – Volume 17, Number 3—March 2011
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Liwang Cui, Department of Entomology, The Pennsylvania State University, 501 ASI Bldg, University Park, PA 16801, USA
Top