Monitoring and Characterization of Oseltamivir-Resistant Pandemic (H1N1) 2009 Virus, Japan, 2009–2010
Makoto Ujike, Miho Ejima, Akane Anraku, Kozue Shimabukuro, Masatsugu Obuchi, Noriko Kishida, Xu Hong, Emi Takashita, Seiichiro Fujisaki, Kazuyo Yamashita, Hiroshi Horikawa, Yumiko Kato, Akio Oguchi, Nobuyuki Fujita, Masato Tashiro, Takato Odagiri

, and the Influenza Virus Surveillance Group of Japan
Author affiliations: Author affiliations: National Institute of Infectious Diseases, Tokyo, Japan (M. Ujike, M. Ejima, A. Anraku, K. Shimabukuro, M. Obuchi, N. Kishida, X. Hong, E. Takashita, S. Fujisaki, K. Yamashita, M. Tashiro, T. Odagiri); National Institute of Technology and Evaluation, Tokyo (H. Horikawa, Y. Kato, A. Oguchi, N. Fujita)
Main Article
Figure 3
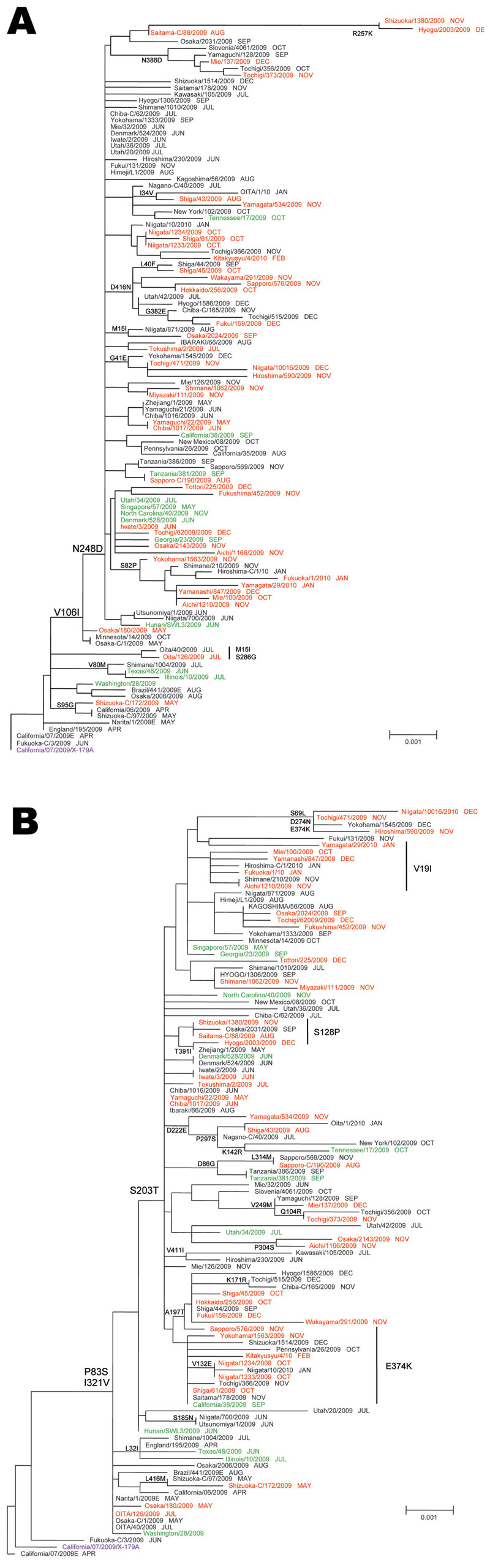
Figure 3. Phylogenetic analysis of influenza pandemic (H1N1) 2009 viruses neuraminidase (NA) (A) and hemagglutinin (HA) genes (B). Most pandemic (H1N1) 2009 viruses possessed the amino acid substitutions S203T in HA and V106I and N248D in NA. Red, oseltamivir-resistant pandemic (H1N1) 2009 from Japan; green, oseltamivir-resistant pandemic (H1N1) 2009 from outside Japan; black, oseltamivir-susceptible (OS) pandemic (H1N1) 2009; purple, 2009–10 current vaccine strains. The sampling month of each isolate is listed following the strain name. The phylogenetic tree of NA and HA genes was constructed by using the neighbor-joining method. Scale bars indicate nucleotide substitutions per site.
Main Article
Page created: July 25, 2011
Page updated: July 25, 2011
Page reviewed: July 25, 2011
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
