Volume 17, Number 9—September 2011
Research
Multiple Reassortment between Pandemic (H1N1) 2009 and Endemic Influenza Viruses in Pigs, United States
Figure 2
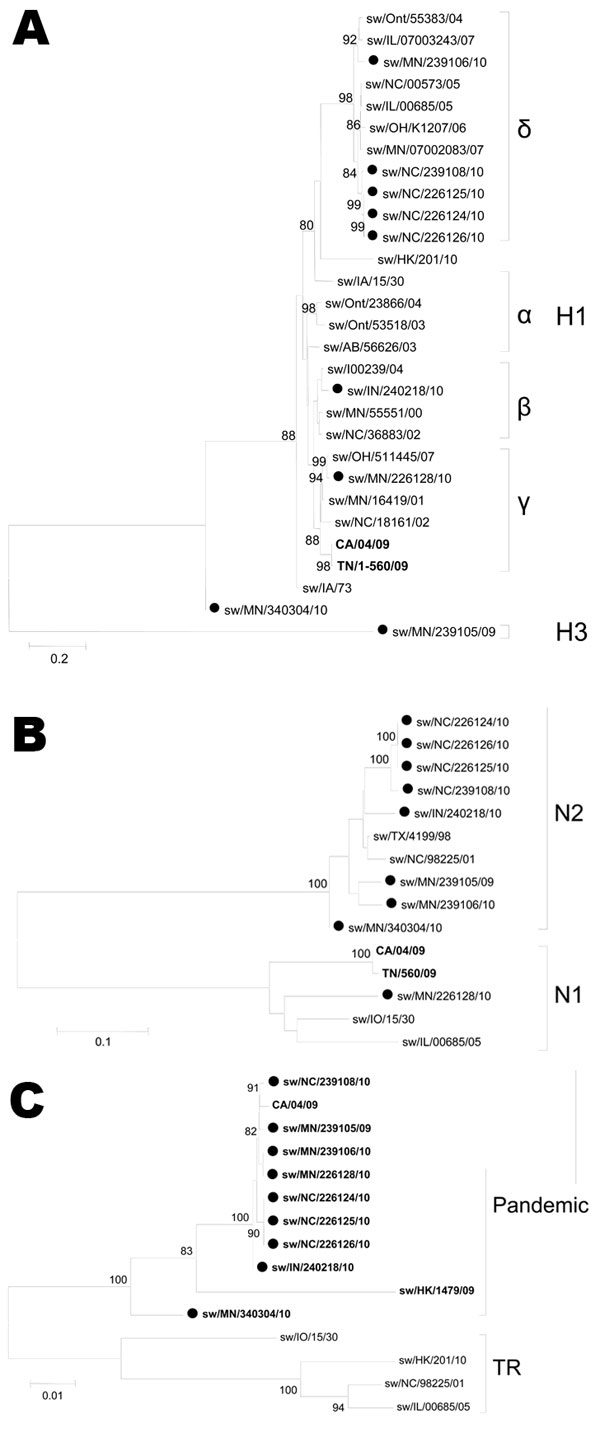
Figure 2. Phylogenetic trees of pandemic reassortant swine influenza viruses compared with currently circulating swine influenza strains: A) hemagglutinin (H); B) neuraminidase (N); C) matrix genes. The trees were constructed by using the neighbor-joining method (Kimura 2-parameter) with 1,000 bootstrap replicates. Only bootstrap values >74 are shown. Swine reassortant strains characterized in this study are indicated with a closed circle. Boldface indicates pandemic segments. Greek letters indicate virus genogroups; α represents classical swine influenza virus and δ seasonal human-like swine influenza virus. TR indicates swine triple reassortant influenza virus. Scale bars indicate nucleotide substitutions per site.
Page created: September 06, 2011
Page updated: September 06, 2011
Page reviewed: September 06, 2011
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.