Volume 18, Number 11—November 2012
Research
HIV Infection and Geographically Bound Transmission of Drug-Resistant Tuberculosis, Argentina
Cite This Article
Citation for Media
Abstract
During 2003–2009, the National Tuberculosis (TB) Laboratory Network in Argentina gave 830 patients a new diagnosis of multidrug-resistant (MDR) TB and 53 a diagnosis of extensively drug- resistant (XDR) TB. HIV co-infection was involved in nearly one third of these cases. Strain genotyping showed that 7 major clusters gathered 56% of patients within restricted geographic areas. The 3 largest clusters corresponded to epidemic MDR TB strains that have been undergoing transmission for >10 years. The indigenous M strain accounted for 29% and 40% of MDR and XDR TB cases, respectively. Drug-resistant TB trends in Argentina are driven by spread of a few strains in hotspots where the rate of HIV infection is high. To curb transmission, the national TB program is focusing stringent interventions in these areas by strengthening infection control in large hospitals and prisons, expediting drug resistance detection, and streamlining information-sharing systems between HIV and TB programs.
During the early 1990s, HIV-associated multidrug-resistant tuberculosis (MDR TB) emerged in Argentina (1). In Buenos Aires, the country’s most heavily populated city, certain multidrug-resistant Mycobacterium tuberculosis strains spread quickly among patients with AIDS (2,3). Specifically, the so-called M strain caused a major MDR TB outbreak at the Hospital Muñiz, a referral treatment center for infectious diseases (4). HIV-infected patients repeatedly seeking assistance at different health centers introduced the M strain into hospitals in nearby districts, where secondary transmission occurred (5). This strain was later responsible for the emergence of MDR TB in HIV-negative patients who had not previously undergone TB treatment (6). In 2002, the M strain was isolated from 2 patients with extensively drug-resistant TB (XDR TB). Two other MDR TB outbreak strains, Ra and Rb, emerged in Rosario, the third largest city in Argentina, simultaneously with the M strain (7).
MDR TB emergence highlighted the need for a MDR/XDR TB surveillance system focused on incidence and transmission. In 2003, the National TB Laboratory Network launched a systematic registry of all incident MDR/XDR TB cases diagnosed throughout the country. The registry includes a genotype database for all MDR/XDR TB patients going back to the initial outbreaks and population studies. We present the findings of a 7-year follow-up study of MDR and XDR TB in Argentina, with emphasis on potential transmission events involving strains responsible for previous outbreaks.
Study Group
Isolates from all patients with newly diagnosed MDR or XDR TB from January 2003 through December 2009 were included in the study (1 isolate per patient, collected at time of diagnosis). MDR TB was defined as disease caused by M. tuberculosis resistant to at least isoniazid and rifampin and XDR TB as disease caused by MDR M. tuberculosis showing further resistance to any fluoroquinolone and any second-line injectable anti-TB drug. A patient with newly diagnosed MDR or XDR TB was defined as a patient with disease first confirmed by drug susceptibility testing (DST) during the study period, regardless of previous treatment history. A hotspot was defined as an area where a MDR TB outbreak had been documented before the study period. Two or more patients were considered to be epidemiologically related when they were in the same place and time or shared similar behavioral risk factors.
Available demographic and clinical data were collected through the national TB laboratory network. A special effort was made to retrieve data from clinical records in special groups, i.e., XDR TB, patients in hotspot areas, and those in clusters with <6 bands in the IS6110 restriction fragment length polymorphism (RFLP). This research was approved by the research review board of the Instituto Nacional de Enfermedades Infecciosas, Administración Nacional de Laboratorios e Institutos de Salud (INEI ANLIS) “Carlos G. Malbran.”
Bacteriologic Studies
On the basis of programmatic guidelines, DST was performed on isolates from patients at risk for drug resistance: patients with TB treatment failure and retreatment, HIV or other concomitant conditions, or exposure to drug-resistant TB in household, prison, or hospital. In Buenos Aires and Rosario, culture and DST are available to test virtually all persons with suspected TB who seek assistance at large referral treatment centers. In the rest of the country, persons not included in the high-risk group are highly unlikely to contract MDR TB. In all, ≈10,000 TB cases are reported annually in Argentina, of which ≈4,500 are diagnosed on the basis of a positive culture. Among cases that are culture-positive, ≈3,000 have isolates submitted for DST; MDR TB is diagnosed for 4% of these patients.
M. tuberculosis DST to first-line drugs (isoniazid, rifampin, streptomycin, ethambutol, and pyrazinamide) was performed in 19 TB network laboratories under regular proficiency testing, according to World Health Organization standards (8). The supranational reference laboratory at Instituto Nacional de Enfermedades Infecciosas ANLIS conducted external quality control, confirmed multidrug resistance, and tested susceptibility to second-line drugs (kanamycin, amikacin, capreomycin, and ofloxacin), according to World Health Organization recommendations (9).
Genotyping
All available isolates underwent standard IS6110 DNA RFLP fingerprinting and spoligotyping (10,11). Patterns were compared by using BioNumerics 5.1 software (Applied Maths, St-Martens-Latem, Belgium), using the Dice coefficient with 1% tolerance and the unweighted pair-group method with arithmetic averages (12). The RFLP pattern of the reference strain M. tuberculosis Mt14323 was used for gel normalization.
For RFLP patterns with >6 bands, a cluster was defined as a group of >2 isolates whose RFLP patterns and spoligotypes were 100% identical when compared with all other patterns found within the study period. RFLP patterns with <6 bands were included in a cluster when epidemiologic links were established in addition to identical spoligotypes. Similarly, a proven epidemiologic link was required for including in a cluster a variant of a cluster genotype; a genotype variant was defined as a genotype with a 1-band difference in the RFLP or 1-spacer difference in the spoligotype but not both. Because of the unusually large size of some clusters, a major cluster was defined as >15 patients and a minor cluster as <15 patients with MDR TB newly diagnosed during the period. Shared International Type (SIT) and genotype family were assigned by consulting the SITVIT database (www.pasteur-guadeloupe.fr:8081/SITVIT) (13), except for genotypes H4, T1-Tuscany, and LAM3/S convergent, which were reclassified as Ural, LAM-Tuscany, and S, respectively, according to Abadia et al. (14). Orphan genotypes were those lacking a SIT in the SITVIT database.
Statistical Analysis
We used univariate and multivariate logistic regression analyses to determine factors associated with being in a cluster and being in a major cluster. The explanatory variables were patient age, country of birth, place of diagnosis, HIV status, previous TB treatment, and disease localization. Within the subgroup included in major clusters, we used logistic regression analysis to determine factors associated with being infected by the M strain. In this latter model, the explanatory variables were patient age, country of birth, HIV status, previous TB treatment, hospital exposure, and isolate drug resistance.
We divided patients into 3 age groups: <15, 16–45, and >45 years of age. Gender was removed from the models because it was associated with particular settings in 2 major clusters; unknown categories were removed from all the variables included in the model. Because of the limited numbers per category, the age category <15 years was removed from the multivariate analyses.
We applied 3 tests to assess the performance of the models: overall model fit, Hosmer & Lemeshow test, and receiver operating characteristic area under the curve. We considered a model to be adequate when values were: overall model fit p<0.2, Hosmer & Lemeshow test p>0.5, and area under the curve >0.70. We used the χ2 test for linear trends for assessing changes in the annual number of MDR TB patients in cluster M compared with changes in numbers in other major clusters. Statistical analyses were performed by using MedCalc version 12 software (MedCalc, Mariakerke, Belgium).
MDR TB Patients, Genotypes, and Clustering
Genotyping Coverage
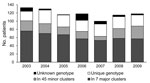
Genotyping was available for isolates from 787/830 (94.8%) newly diagnosed MDR TB patients registered during the study period (2003, 93.2%; 2004, 97.7%; 2005, 99.1%; 2006, 86.4%; 2007, 93.0%; 2008, 96.5%; 2009, 97.5%) (Figure 1). Coverage was lower in 2006 because of a technical mishap that resulted in a loss in the isolate collection.
Genotype Family Distribution
The 3 predominant genotype families were LAM (38.8%), Haarlem (36.3%), and T (13.9%). Other genotypes were S (2.8%), U (1.7%), Beijing (1.5%), X (0.9%), and Ural (0.4%); orphan genotypes accounted for 3.8%. Within the 3 predominant genotype families, the most frequent subfamilies were H2 (29.5%), LAM3 (16.4%), T1 (8.9%), LAM5 (6.6%), LAM9 (6.5%), and Tuscany (5.5%). Of 12 patients carrying Beijing genotypes, 1 was born in Indonesia, 7 in Peru, and 4 in South America with no information on country of birth.
Clusters
Of 787 patients for whom isolate genotype was available, 438 (55.7%) fitted into 7 major clusters and 151 (19.2%) into 45 minor clusters; 198 (25.2%) harbored unique genotypes (Table 1). In the multivariate regression analysis, the outcome of being in a cluster was significantly dependent on being 15–44 years of age, born in Argentina, and HIV-infected. Being in a major cluster was significantly dependent on the 2 latter predictors and of having the MDR TB diagnosis occur in a hotspot area (Table 2).
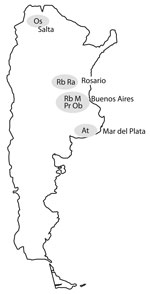
Characteristics of the 7 major clusters are described in Table 3 and DST profiles in Table 4. Genotype patterns are shown in Figure 2 and geographic distribution in Figure 3. Altogether, the 3 largest clusters (M, Ra, and Rb) accounted for 355 (45.1%) patients; isolate patterns matched 3 genotypes previously associated with MDR TB outbreaks (4,7,15). The other 4 major clusters (Pr, At, Ob, and Os) accounted for 83 (10.5%) patients; these genotypes had been reported only sporadically before the study period.
The predominant cluster, M, was largely confined to the city of Buenos Aires and the surrounding area, with only 5/228 patients having MDR TB diagnosed elsewhere. Twenty patients in this cluster were immigrants from neighboring countries (Bolivia 11, Paraguay 6, Peru 2, Uruguay 1). Most patients had >1 commonly acknowledged risk factors for MDR TB (129 patients had 1, 64 had 2, and 16 had 3 risk factors) (Table 3). The cluster included the 2 previously reported outbreak variants of the M strain (4), Mm in 180 patients and Mn in 35 patients (Figure 2), and 9 sporadic variants, observed in 13 patients who had proven epidemiologic links with other patients in cluster M. An isolate resistant to 5 drugs was strongly associated with disease produced by the M strain (Table 5). The numbers of patients affected by this strain decreased significantly within the period when compared with the numbers of patients in the other 6 major clusters (p = 0.002). In particular, the proportion of HIV-infected patients affected by the M strain decreased significantly during the study period, from 65% in 2003 to 24% in 2009 (p = 0.02; Figure 4). No similar trend was observed in the HIV-negative group (p = 0.77).
The second major cluster, Ra, was mainly limited to the overpopulated area of Rosario City and surroundings, another MDR TB hotspot (7). Only 8 patients in this cluster were found outside that area. Of 38 patients in the third major cluster, Rb, 26 received a diagnosis of MDR TB in Buenos Aires, including 10 transvestite sex workers; 8 patients had the disease diagnosed in Rosario, and 4 elsewhere. Cluster Pr consisted mainly of inmates in several state prisons for men. An association of gender with clustering was only observed in clusters Rb and Pr, in which men predominated (32/38 and 24/26, respectively).
XDR TB Patients and Genotypes
XDR TB was newly diagnosed in 53 patients during 2003–2009. Of these patients, 37 first received a diagnosis of MDR TB during the same period, so these patients were included in the MDR and XDR TB groups. The other 16 XDR TB patients received a diagnosis of MDR TB before this period. The male:female ratio for XDR TB patients was 1.25:1; median age was 37.4 years (SD 11.6, range 21–72 years). Fifty-two patients were born in South America (Argentina 30, Bolivia 4, Peru 6, Paraguay 1, Brazil 1, undetermined 11), and 1 was born in Indonesia.
Characteristics of XDR TB patients related to clustering are described in Table 6. Four major clusters (M, Os, At, and Rb) comprised 31 (58%) XDR TB patients; 6/14 HIV-positive patients with XDR TB were in cluster Os, and 5 in cluster M. Five of 11 immigrants from other South American countries to Argentina harbored the M strain, and the patient from Indonesia harbored a Beijing strain. Of 15 patients with no record of previous TB treatment, 12 were included in major clusters. Annual numbers of XDR TB patients by genotype are shown in Table 7.
The annual number of newly diagnosed MDR and XDR TB cases decreased slightly, with minor fluctuations, during the study period. HIV infection was associated with almost one third of MDR TB cases; this proportion is 2–4× higher than that attributed by different studies to all forms of TB in the country (16,17). As previously observed (18), annual fluctuations in the numbers of total MDR/XDR TB cases during the study period paralleled closely annual fluctuations in the numbers of HIV-infected patients.
All 7 major clusters in our study were connected with particular geographic areas, institutional settings, or both. Furthermore, most patients in these clusters underwent TB treatment in health centers that had ongoing MDR TB transmission or had a household or a prison contact with persons who had MDR TB. These findings indicate that these major clusters represent true transmission events.
Many cases in this outbreak were caused by the M strain, an apparently autochthonous outbreak genotype. In a countrywide survey performed in 1998, this strain was found to be responsible for 42% of all MDR TB cases but was confined to the metropolitan area of Buenos Aires (National TB Laboratory Network, unpub. data). Since then, the M strain has been the most frequently identified in every MDR TB investigation performed in the country. We found the M strain to be the most prevalent and that its transmission was virtually restricted to the area of the initial outbreak. However, its numbers decreased by more than half within the study period, particularly among HIV-infected patients, which suggests that the epidemic curve of the M strain has entered a declining phase. The other 2 strains associated with previous MDR TB outbreaks, Ra and Rb, were found to persist with lower, fluctuating frequencies.
The expansion of M strain transmission in Argentina was initially fostered by clinical mismanagement. During the early 1990s, patients with advanced AIDS were hospitalized in large, referral treatment centers, where they shared facilities with patients who had MDR TB. At that time, virtually no respiratory protection policy was in force because it was wrongly assumed that MDR TB patients were barely infectious. After genotyping confirmed outbreaks of MDR TB among the patients with AIDS (4), hospital infection control interventions were adopted, microbiologic diagnosis and drug-resistance detection were expedited, and second-line TB drugs and highly active antiretroviral therapy became available. As a result of those interventions, hospital transmission was substantially reduced but not completely controlled (18). At the time of our study, however, the M strain had long expanded beyond the hospital environment (6).
The national M. tuberculosis genotype database in Argentina identified very few patients with non–MDR TB harboring H2 genotypes (V. Ritacco, unpub. data). Another study supported this observation (19), and the H2 genotype was found to be infrequent in other South American countries (13,20–22). The IS6110 RFLP pattern of the M strain was absent outside Argentina in the Ibero-American MDR TB genotype database (23) and was not present in the M. tuberculosis genotype database in the Netherlands (D. van Soolingen, pers. comm.). This pattern was registered in other countries, anecdotally, in 2 MDR TB patients with AIDS: a patient from Argentina who died in San Francisco, California, shortly after his arrival in the United States (24); and a patient from Asuncion, Paraguay, who visited the Hospital Muñiz in Buenos Aires each month for antiretroviral therapy (20).
Different M. tuberculosis genotypes may have affinity with certain geographic areas and ethnic groups (25), which could explain why the M strain persists in the Buenos Aires area. In addition, most persons at risk for MDR TB are underprivileged and cannot afford to travel far distances. This mobility limitation might also help to explain why this strain has remained virtually confined to the original hotspot area.
A small number of patients affected by the M strain were immigrants from neighboring countries who had settled in Buenos Aires. Cross-border and domestic migration toward large metropolitan areas is a long-observed demographic and public health concern in Argentina. More than 80% of the patients in this study were assisted in metropolises designated as MDR TB hotspots. Even though immigrants with TB have access to higher quality health care in these areas than in other parts of the country, they are also at higher risk of becoming newly infected with an outbreak MDR TB strain.
The M strain was overrepresented among patients with XDR TB; isolate resistance to >5 anti-TB drugs was found to be a strong predictor of disease caused by the M strain (4,26). The accumulation of drug resistance–conferring mutations would be expected to have reduced the epidemiologic fitness of this strain, but it has prevailed for 15 years. The epidemiologic fitness of a strain can be influenced by a range of factors, e.g., the genetic backgrounds of host and pathogen, host–pathogen interactions, and the environment (27–29). Compensatory evolution restoring in vivo fitness, as well as social and behavioral factors, might have played a role in the epidemiologic persistence of the M strain (30). These factors might also have preferentially fostered the spread of drug-resistant strains of the H2 genotype in our setting. Further studies are needed to evaluate the most critical risk factors.
Drug-resistance profiles were not uniform within the M strain clusters. The variations in susceptibility to individual drugs reflect the existence of various ongoing chains of transmission, some of which might have started before, or simultaneously with, the first documented outbreak. One limitation of our study is the failure to identify individual chains. Factors that precluded the reliable characterization of subclusters were the long time elapsed since the outbreak onset, the insufficient epidemiologic documentation in many cases, and the unavailability of additional molecular markers.
Our study has another major limitation. Incomplete demographic and clinical data on patients were retrieved, and several observations had missing values. If missing values were systematically associated with a given force or factor, results presented here would be biased. We are not aware of any association of missing values with the dependent variables under study and assume that those data were missing at random. Missing values may have affected the analyses by reducing the number of observations, which may have reduced the power of the model to detect significant associations but without necessarily biasing the associations reported. However, the possibility that bias might have resulted from missing data cannot be ruled out. Therefore, statistical significances of our analyses should be interpreted cautiously.
In the MDR TB hotspots in Argentina, the distinction between primary and acquired MDR TB on the basis of a history of previous TB treatment was not decisive because patients could have been exposed to hospital-associated MDR TB infection while being treated for community-acquired TB. This fact could explain why clustering was not more frequent among patients without previous TB treatment in our study.
The national TB network includes all the laboratories performing bacteriological TB diagnosis in the country; therefore, the patients in this study represent all newly diagnosed MDR TB cases in Argentina. The structure, geographic coverage, and personnel of the TB laboratory network are adequate to provide DST for all patients at risk for MDR TB in Argentina. However, a few MDR TB patients might remain undiagnosed because of operational factors, e.g., inefficient detection of risk factors, insufficient or delayed requests for DST, and disorganized information systems.
The geographically restricted distribution of successful MDR TB genotypes that we found has public health implications. As a result of this study, specific interventions are being reinforced, particularly in the MDR TB hotspots: implementing universal culture and strategies to expedite drug resistance detection; decentralizing specialized health care; streamlining information-sharing systems between HIV and TB programs; and strengthening administrative infection control measures in prisons and large hospitals with high TB infection load. A national advisory group on MDR TB clinical management has also been recently created. Control interventions have already started to reduce MDR TB spread in the hospital that was the epicenter of the main outbreak (17). Still, centrally coordinated actions are needed in Argentina to curb long-term transmission of MDR TB.
Dr Ritacco is a scientist at the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), Argentina. Her main research interests are molecular epidemiology and molecular diagnosis of tuberculosis.
Acknowledgments
On behalf of TB laboratory network, the following persons contributed substantially to this work: D. Ballester, L. Casimir, M. Gutiérrez, M. Hoffman, S. Kaufman, N. Kempf, G. Ojea, A. Procopio (Buenos Aires City); M.I. Anganuzzi, V. Delle Donne, T. Hernández, A. Mariñansky, N. Morcillo, S. Morla, A. Reniero, L. Rivas, A. Rossetti, A. Togneri, A. Zapata (Buenos Aires Province); M.I. Gigli, C. Pellegrini (Santa Fe); M. Cisneros (Salta); M.C. Cosiansi, L. Wolff (Córdoba); G. Bichara (Tucumán); M. Pacce (Corrientes), V. Carrió (San Juan); M. Fantín (Formosa); H. Rousselle (Mendoza); R. Piloni (Misiones); G. Torres (Neuquén), M. Gunia (Chaco); E. Fernández (Chubut); L. Casali (Catamarca); O. Belforte (Santa Cruz); N. Blázquez (Río Negro); and G. Jacquier (Sistema Penitenciario).
We thank Andrés M. Pérez for valuable advice on statistical analysis.
This work was supported by FonCyT, Argentina (project PAE-PICT2323), and the EC under contract HEALTH-F3_2007-201690 (FAST-XDR-DETECT).
References
- de Kantor IN, Ritacco V, Barrera L. Is multidrug-resistant tuberculosis an emergent infection in Buenos Aires? [in Spanish]. Medicina (B Aires). 1996;56:102–4.PubMedGoogle Scholar
- Ritacco V, Reniero A, Ambroggi M, Barrera LB, López B, Di Lonardo M, Inter-hospital spread of multidrug-resistant tuberculosis in Buenos Aires, Argentina. Tuber Lung Dis. 1996;77(Suppl 2):63.
- Morcillo N, Alito A, Romano MI, Cataldi A, Dolmann A, Reniero A, Multidrug resistant tuberculosis outbreak in Buenos Aires. DNA fingerprinting analysis of isolates. Medicina (B Aires). 1996;56:45–7.PubMedGoogle Scholar
- Ritacco V, Di Lonardo M, Reniero A, Ambroggi M, Barrera L, Dambrosi A, Nosocomial spread of Human Immunodeficiency Virus-related multidrug-resistant tuberculosis in Buenos Aires. J Infect Dis. 1997;176:637–42. DOIPubMedGoogle Scholar
- Poggio G, Togneri A, Reniero A, Insúa A, Guerra R, Dinerstein E, AIDS-related multidrug resistant tuberculosis “M” strain spreads within two hospitals in Buenos Aires suburbs. Int J Tuberc Lung Dis. 1997;1(Supp1):523.PubMedGoogle Scholar
- Palmero D, Ritacco V, Ambroggi M, Marcela N, Barrera L, Capone L, Multidrug-resistant tuberculosis in HIV-negative patients, Buenos Aires, Argentina. Emerg Infect Dis. 2003;9:965–9. DOIPubMedGoogle Scholar
- Aita J, Barrera L, Reniero A, López B, Biglione J, Weisburd G, Hospital transmission of multidrug-resistant Mycobacterium tuberculosis in Rosario, Argentina. Medicina (B Aires). 1996;56:48–50.PubMedGoogle Scholar
- World Health Organization. Guidelines for surveillance of drug resistance in tuberculosis. WHO/HTM/TB/2009.442. Geneva: The Organization; 2009.
- World Health Organization. Policy guidance on TB drug susceptibility testing (DST) of second-line drugs. WHO/HTM/TB/2008.392. Geneva: The Organization; 2008.
- van Embden JDA, Cave MD, Crawford JT, Dale JW, Eisenach KD, Gicquel B, Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993;31:406–9.PubMedGoogle Scholar
- Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35:907–14.PubMedGoogle Scholar
- Heersma HF, Kremer K, van Embden J. Computer analysis of IS6110 RFLP patterns of Mycobacterium tuberculosis. Methods Mol Biol. 1998;101:395–422.PubMedGoogle Scholar
- Brudey K, Driscoll JR, Rigouts L, Prodinger WM, Gori A, Al-Hajoj SA, Mycobacterium tuberculosis complex genetic diversity: mining the fourth international spoligotyping database (SpolDB4) for classification, population genetics and epidemiology. BMC Microbiol. 2006;6:23. DOIPubMedGoogle Scholar
- Abadia E, Zhang J, dos Vultos T, Ritacco V, Kremer K, Aktas E, Resolving lineage assignation on Mycobacterium tuberculosis clinical isolates classified by spoligotyping with a new high-throughput 3R SNPs based method. Infect Genet Evol. 2010;10:1066–74. DOIPubMedGoogle Scholar
- Palmero D, Ritacco V, Ruano S, Ambroggi M, Cusmano L, Romano M, Multidrug-resistant tuberculosis outbreak in transvestite sex workers, Buenos Aires, Argentina. Int J Tuberc Lung Dis. 2005;9:1168–70.PubMedGoogle Scholar
- Pando MA, De Salvo C, Bautista CT, Eyzaguirre L, Carrion G, Feola M, Human immunodeficiency virus and tuberculosis in Argentina: prevalence, genotypes and risk factors. J Med Microbiol. 2008;57:190–7. DOIPubMedGoogle Scholar
- Ritacco V, Morgado M, Garcia LF. The situation of HIV/M. tuberculosis in South America. Open Infect Dis J. 2011, 5, (Suppl 1–M7) 81–88 [cited May 20, 2012]. http://benthamscience.com/open/toidj/articles/V005/SI0013TOIDJ/81TOIDJ.htm
- Waisman JL, Palmero DJ, Güemes-Gurtubay JL, Videla JJ, Moretti B, Cantero M, Evaluation of the control measures adopted against an epidemic of AIDS-related multidrug-resistant tuberculosis in a Latin-American hospital [in Spanish]. Enferm Infecc Microbiol Clin. 2006;24:71–6. DOIPubMedGoogle Scholar
- Gonzalo X, Ambroggi M, Cordova E, Brown T, Poggi S, Drobniewski F. Molecular epidemiology of Mycobacterium tuberculosis, Buenos Aires, Argentina. Emerg Infect Dis. 2011;17:528–31. DOIPubMedGoogle Scholar
- Candia N, Lopez B, Zozio T, Carrivale M, Diaz C, Russomando G, First insight into Mycobacterium tuberculosis genetic diversity in Paraguay. BMC Microbiol. 2007;7:75. DOIPubMedGoogle Scholar
- Aristimuño L, Armengol R, Cebollada A, España M, Guilarte A, Lafoz C, Molecular characterisation of Mycobacterium tuberculosis isolates in the First National Survey of Anti-Tuberculosis Drug Resistance from Venezuela. BMC Microbiol. 2006;6:90. DOIPubMedGoogle Scholar
- Lazzarini LC, Huard RC, Boechat NL, Gomes HM, Oelemann MC, Kurepina N, Discovery of a novel Mycobacterium tuberculosis lineage that is a major cause of tuberculosis in Rio de Janeiro, Brazil. J Clin Microbiol. 2007;45:3891–902. DOIPubMedGoogle Scholar
- Ritacco V, Iglesias MJ, Ferrazoli L, Monteserin J, Dalla Costa ER, Cebollada A, Conspicuous multidrug-resistant Mycobacterium tuberculosis cluster strains do not trespass country borders in Latin America and Spain. Infect Genet Evol. 2012;12:711–7. DOIPubMedGoogle Scholar
- Burgos MV, Méndez JC, Ribon W. Molecular epidemiology of tuberculosis: methodology and applications. Biomedica. 2004;24(Supp1):188–201.PubMedGoogle Scholar
- Gagneux S, DeRiemer K, Van T, Kato-Maeda M, de Jong BC, Narayanan S, Variable host-pathogen compatibility in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2006;103:2869–73. DOIPubMedGoogle Scholar
- Palmero D, Ritacco V, Ambroggi M, Poggi S, Güemes Gurtubay J, Alberti F, Multidrug-resistant tuberculosis in AIDS patients at the beginning of the millennium. Medicina (B Aires). 2006;66:399–404.PubMedGoogle Scholar
- Gagneux S, Long CD, Small PM, Van T, Schoolnik GK, Bohannan BJM. The competitive cost of antibiotic resistance in Mycobacterium tuberculosis. Science. 2006;312:1944–6. DOIPubMedGoogle Scholar
- Luciani F, Sisson SA, Jiang H, Francis AR, Tanaka MM. The epidemiological fitness cost of drug resistance in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2009;106:14711–5. DOIPubMedGoogle Scholar
- O’Sullivan DM, McHugh TD, Gillespie SH. Mapping the fitness of Mycobacterium tuberculosis strains: a complex picture. J Med Microbiol. 2010;59:1533–5. DOIPubMedGoogle Scholar
- Comas I, Borrell S, Roetzer A, Rose G, Malla B, Kato-Maeda M, Whole-genome sequencing of rifampicin-resistant Mycobacterium tuberculosis strains identifies compensatory mutations in RNA polymerase genes. Nat Genet. 2011;44:106–10. DOIPubMedGoogle Scholar
Figures
Tables
Cite This Article1Additional members of the National TB Laboratory Network who contributed data are listed at the end of this article.
Table of Contents – Volume 18, Number 11—November 2012
| EID Search Options |
|---|
|
|
|
|
|
|


Please use the form below to submit correspondence to the authors or contact them at the following address:
Viviana Ritacco, INEI ANLIS Carlos Malbrán, Vélez Sarsfield 563, (1281) Buenos Aires, Argentina
Top