Volume 18, Number 3—March 2012
Research
A Systematic Approach for Discovering Novel, Clinically Relevant Bacteria
Figure 2
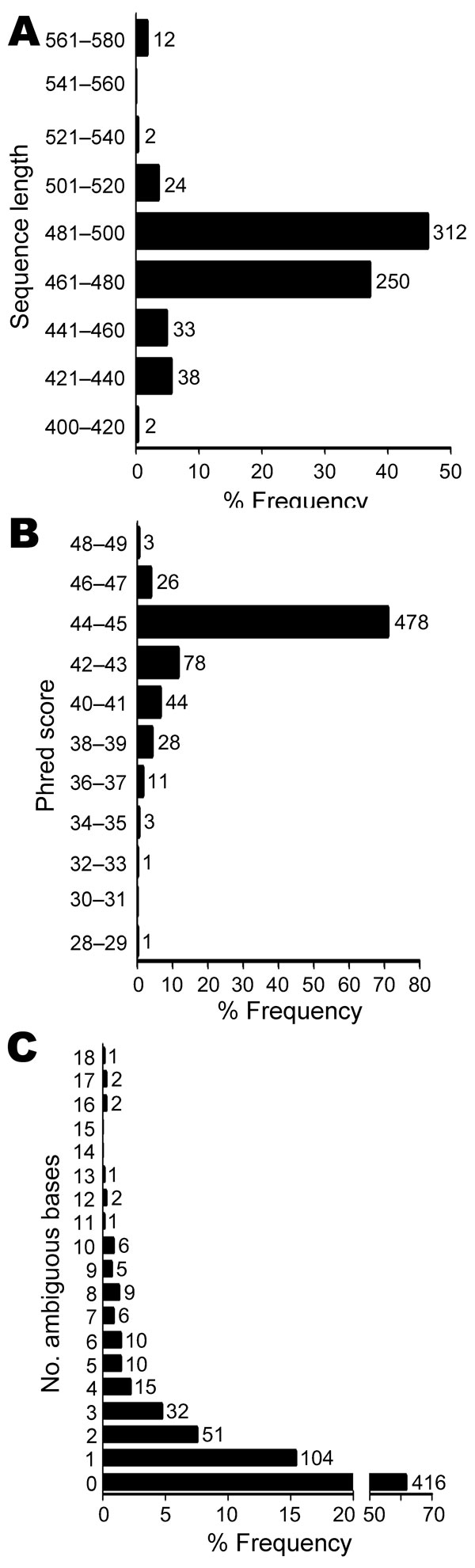
Figure 2. Sequence quality and number of ambiguous bases for 673 unidentified bacterial isolates. The median sequence length was 480 bases, with 84% of sequences in the range of 461 to 500 bases (A). The median phred sequence quality score was 45 (B). Most sequences had no ambiguous positions (n = 416, 61.8%). Up to 18 ambiguous positions were seen in isolates with multiple, nonidentical copies of the 16S rRNA gene (C). The x-axes indicate relative frequency. Numbers above columns represent isolate counts.
1These authors contributed equally to this article.
Page created: February 16, 2012
Page updated: February 16, 2012
Page reviewed: February 16, 2012
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.