Volume 18, Number 4—April 2012
Research
Identification of Intermediate in Evolutionary Model of Enterohemorrhagic Escherichia coli O157
Figure
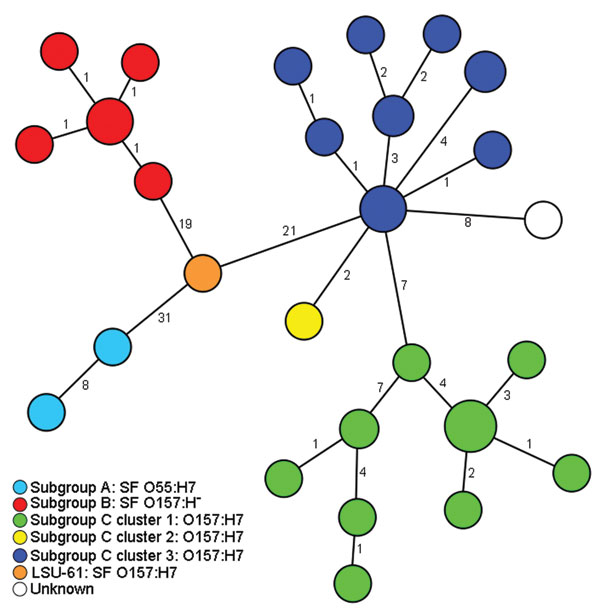
Figure. Minimum-spanning tree based on single-nucleotide polymorphism (SNP) genotypes illustrating the phylogeny of 50 enterohemorrhagic Escherichia coli O157:H7/H– and O55:H7 isolates and the intermediate position of strain LSU-61 during the evolution of O157. Each node represents a unique SNP genotype. The size of each node is proportional to the number of isolates per SNP genotype based on sequence analysis of 51,041 bp comprising 92 partial open reading frames. Numbers on lines between nodes represent distances between the nodes, i.e., the number of SNPs. The node size is proportional to the number of strains sharing the same genotype. Strains are colored according to their classification into subgroups and clusters based on information from Saikh and Tarr (11) and Leopold et al. (12). Strain LSU-61 represents a potential intermediate interlinking all 3 subgroups. SF, sorbitol fermenting.
References
- Holtz LR, Neill MA, Tarr PI. Acute bloody diarrhea: a medical emergency for patients of all ages. Gastroenterology. 2009;136:1887–98. DOIPubMedGoogle Scholar
- Levine MM. Escherichia coli that cause diarrhea: enterotoxigenic, enteropathogenic, enteroinvasive, enterohemorrhagic, and enteroadherent. J Infect Dis. 1987;155:377–89. DOIPubMedGoogle Scholar
- Tarr PI, Gordon CA, Chandler WL. Shiga toxin–producing Escherichia coli and haemolytic uraemic syndrome. Lancet. 2005;365:1073–86. DOIPubMedGoogle Scholar
- Karch H, Tarr PI, Bielaszewska M. Enterohaemorrhagic Escherichia coli in human medicine. Int J Med Microbiol. 2005;295:405–18. DOIPubMedGoogle Scholar
- Karch H, Bielaszewska M. Sorbitol-fermenting Shiga toxin–producing Escherichia coli O157:H– strains: epidemiology, phenotypic and molecular characteristics, and microbiological diagnosis. J Clin Microbiol. 2001;39:2043–9. DOIPubMedGoogle Scholar
- Karch H, Mellmann A, Bielaszewska M. Epidemiology and pathogenesis of enterohaemorrhagic Escherichia coli. Berl Munch Tierarztl Wochenschr. 2009;122:417–24.PubMedGoogle Scholar
- Werber D, Bielaszewska M, Frank C, Stark K, Karch H. Watch out for the even eviler cousin—sorbitol-fermenting E coli O157. Lancet. 2011;377:298–9. DOIPubMedGoogle Scholar
- Feng P, Lampel KA, Karch H, Whittam TS. Genotypic and phenotypic changes in the emergence of Escherichia coli O157:H7. J Infect Dis. 1998;177:1750–3. DOIPubMedGoogle Scholar
- Feng PCH, Monday SR, Lacher DW, Allison L, Siitonen A, Keys C, Genetic diversity among clonal lineages within Escherichia coli O157:H7 stepwise evolutionary model. Emerg Infect Dis. 2007;13:1701–6.PubMedGoogle Scholar
- Shaikh N, Tarr PI. Escherichia coli O157:H7 Shiga toxin–encoding bacteriophages: integrations, excisions, truncations, and evolutionary implications. J Bacteriol. 2003;185:3596–605. DOIPubMedGoogle Scholar
- Leopold SR, Magrini V, Holt NJ, Shaikh N, Mardis ER, Cagno J, A precise reconstruction of the emergence and constrained radiations of Escherichia coli O157 portrayed by backbone concatenomic analysis. Proc Natl Acad Sci U S A. 2009;106:8713–8.PubMedGoogle Scholar
- Dunn JR, Keen JE, Moreland D, Alex T. Prevalence of Escherichia coli O157:H7 in white-tailed deer from Louisiana. J Wildl Dis. 2004;40:361–5.PubMedGoogle Scholar
- Perna NT, Plunkett G III, Burland V, Mau B, Glasner JD, Rose DJ, Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001;409:529–33. DOIPubMedGoogle Scholar
- Shaikh N, Holt NJ, Johnson JR, Tarr PI. Fim operon variation in the emergence of enterohemorrhagic Escherichia coli: an evolutionary and functional analysis. FEMS Microbiol Lett. 2007;273:58–63. DOIPubMedGoogle Scholar
- Karch H, Wiss R, Gloning H, Emmrich P, Aleksic S, Bockemühl J. Hemolytic-uremic syndrome in infants due to verotoxin-producing Escherichia coli [in German]. Dtsch Med Wochenschr. 1990;115:489–95. DOIPubMedGoogle Scholar
- Kulasekara BR, Jacobs M, Zhou Y, Wu Z, Sims E, Saenphimmachak C, Analysis of the genome of the Escherichia coli O157:H7 2006 spinach-associated outbreak isolate indicates candidate genes that may enhance virulence. Infect Immun. 2009;77:3713–21. DOIPubMedGoogle Scholar
- Hayashi T, Makino K, Ohnishi M, Kurokawa K, Ishii K, Yokoyama K, Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 2001;8:11–22. DOIPubMedGoogle Scholar
- Jenke C, Harmsen D, Weniger T, Rothgänger J, Hyytiä-Trees E, Bielaszewska M, Phylogenetic analysis of enterohemorrhagic Escherichia coli O157, Germany, 1987–2008. Emerg Infect Dis. 2010;16:610–6.PubMedGoogle Scholar
- Friedrich AW, Bielaszewska M, Zhang W, Pulz M, Kuczius T, Ammon A, Escherichia coli harboring Shiga toxin 2 gene variants: frequency and association with clinical symptoms. J Infect Dis. 2002;185:74–84. DOIPubMedGoogle Scholar
- Mellmann A, Bielaszewska M, Zimmerhackl LB, Prager R, Harmsen D, Tschäpe H, Enterohemorrhagic Escherichia coli in human infection: in vivo evolution of a bacterial pathogen. Clin Infect Dis. 2005;41:785–92. DOIPubMedGoogle Scholar
- Prager R, Strutz U, Fruth A, Tschäpe H. Subtyping of pathogenic Escherichia coli strains using flagellar (H)-antigens: serotyping versus fliC polymorphisms. Int J Med Microbiol. 2003;292:477–86. DOIPubMedGoogle Scholar
- Sonntag AK, Prager R, Bielaszewska M, Zhang W, Fruth A, Tschäpe H, Phenotypic and genotypic analyses of enterohemorrhagic Escherichia coli O145 strains from patients in Germany. J Clin Microbiol. 2004;42:954–62. DOIPubMedGoogle Scholar
- Zhang Y, Laing C, Steele M, Ziebell K, Johnson R, Benson AK, Genome evolution in major Escherichia coli O157:H7 lineages. BMC Genomics. 2007;8:121. DOIPubMedGoogle Scholar
- Wilson K. Preparation of genomic DNA from bacteria. Curr Protoc Mol Biol. 2001;Chapter 2:Unit 2.4.
- Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol. 2006;60:1136–51. DOIPubMedGoogle Scholar
- Dugan KA, Lawrence HS, Hares DR, Fisher CL, Budowle B. An improved method for post-PCR purification for mtDNA sequence analysis. J Forensic Sci. 2002;47:811–8.PubMedGoogle Scholar
- Rump LV, Strain EA, Cao G, Allard MW, Fischer M, Brown EW, Draft genome sequences of six Escherichia coli isolates from the stepwise model emergence of Escherichia coli O157:H7. J Bacteriol. 2011;193:2058–9. DOIPubMedGoogle Scholar
- Besser TE, Shaikh N, Holt NJ, Tarr PI, Konkel ME, Malik-Kale P, Greater diversity of Shiga toxin–encoding bacteriophage insertion sites among Escherichia coli O157:H7 isolates from cattle than in those from humans. Appl Environ Microbiol. 2007;73:671–9. DOIPubMedGoogle Scholar
- Bielaszewska M, Prager R, Zhang W, Friedrich AW, Mellmann A, Tschäpe H, Chromosomal dynamism in progeny of outbreak-related sorbitol-fermenting enterohemorrhagic Escherichia coli O157:NM. Appl Environ Microbiol. 2006;72:1900–9. DOIPubMedGoogle Scholar
- García-Sánchez A, Sanchez S, Rubio R, Pereira G, Alonso JM, Hermoso de Mendoza J, Presence of Shiga toxin–producing E. coli O157:H7 in a survey of wild artiodactyls. Vet Microbiol. 2007;121:373–7. DOIPubMedGoogle Scholar
- Díaz S, Vidal D, Herrera-Leon S, Sanchez S. Sorbitol-fermenting, β-glucuronidase–positive, Shiga toxin–negative Escherichia coli O157:H7 in free-ranging red deer in south-central Spain. Foodborne Pathog Dis. 2011;8:1313–5. DOIPubMedGoogle Scholar
- Lacher DW, Steinsland H, Blank TE, Donnenberg MS, Whittam TS. Molecular evolution of typical enteropathogenic Escherichia coli: clonal analysis by multilocus sequence typing and virulence gene allelic profiling. J Bacteriol. 2007;189:342–50. DOIPubMedGoogle Scholar
- Bielaszewska M, Köck R, Friedrich AW, von Eiff C, Zimmerhackl LB, Karch H, Shiga toxin–mediated hemolytic uremic syndrome: time to change the diagnostic paradigm? PLoS ONE. 2007;2:e1024. DOIPubMedGoogle Scholar
- Mellmann A, Lu S, Karch H, Xu J, Harmsen D, Schmidt MA, Recycling of Shiga toxin 2 genes in sorbitol-fermenting enterohemorrhagic Escherichia coli O157:NM. Appl Environ Microbiol. 2008;74:67–72. DOIPubMedGoogle Scholar
- Manning SD, Motiwala AS, Springman AC, Qi W, Lacher DW, Ouellette LM, Variation in virulence among clades of Escherichia coli O157:H7 associated with disease outbreaks. Proc Natl Acad Sci U S A. 2008;105:4868–73. DOIPubMedGoogle Scholar
- Zhang W, Qi W, Albert TJ, Motiwala AS, Alland D, Hyytiä-Trees EK, Probing genomic diversity and evolution of Escherichia coli O157 by single nucleotide polymorphisms. Genome Res. 2006;16:757–67. DOIPubMedGoogle Scholar