Volume 18, Number 7—July 2012
Research
Enterococcus faecalis Clones in Poultry and in Humans with Urinary Tract Infections, Vietnam
Cite This Article
Citation for Media
Abstract
Enterococcus spp. as pathogens have increased, but the sources of infection often remain unclear. To investigate whether poultry might be a reservoir for E. faecalis–associated urinary tract infections (UTIs) in humans, we characterized E. faecalis isolates from patients in Vietnam with UTIs during January 2008–January 2010 and poultry living in close contact with them by multilocus sequence typing (MLST), pulsed-field gel electrophoresis, analysis of antimicrobial drug susceptibility patterns, and sequencing of virulence genes. In 7 (23%) of 31 UTI cases, we detected identical MLST, indistinguishable or closely related pulsed-field gel electrophoresis patterns, and similar antimicrobial drug susceptibility patterns. Isolates from urine and poultry showed identical virulence gene profiles, except for 1 variation, and individual genes showed identical sequences. The homology of isolates from urine and poultry further indicates the zoonotic potential and global spread of E. faecalis sequence type 16, which recently was reported in humans with endocarditis and in pigs in Denmark.
Enterococci are commensals of the human and animal gastrointestinal tract and opportunistic pathogens that cause urinary tract infections (UTIs), endocarditis, and sepsis (1). Nosocomial infections caused by enterococci have increased; these pathogens are now the third most common at hospitals after Escherichia coli and Staphylococcus aureus (2); and enterococci are frequently recorded as the cause of UTIs, wound infections, bacteremia, and endocarditis (3–6).
The sources of enterococcal infections in humans are not clear, but animal reservoirs have been suggested (2,4,7–9). A study comparing enterococcal isolates from 4 European countries and the United States demonstrated that E. faecalis isolated from pigs in Portugal had pulsed-field gel electrophoresis (PFGE) patterns identical to those of multidrug-resistant isolates at hospitals in Spain, Italy, and Portugal, all of which were shown by multilocus sequence typing (MLST) to belong to sequence type (ST) 6 (7). In Denmark, high-level gentamicin-resistant E. faecalis of ST16 with an identical PFGE pattern was isolated from pigs and from humans with endocarditis (9). Identical and closely related PFGE patterns were demonstrated by isolates from humans and from pork and chicken meat in the United States, all of which contained high-level gentamicin-resistant genes (4). Our objective was to characterize epidemiologically related E. faecalis isolated from humans with UTIs and from poultry living in the same households in Vietnam to evaluate the zoonotic potential of E. faecalis.
Recruitment of Patients, Urine Collection, and Bacterial Culture of Urine
Urine samples were collected during January 2008–January 2010 at the Military Medical University, Hospital 103, in Ha Dong, Hanoi. Patients with clinical symptoms of UTI (i.e., >1 of the following symptoms: frequent urination; painful urination; hematuria; cloudy urine; or pain in pelvic area, flank, or low back) were referred from nearby pharmacies and informed about the project. A midstream urine sample was collected at the hospital under supervision of a nurse. Only patients with uncomplicated UTIs were included; patients reporting underlying diseases, such as hematologic disorders, respiratory infections, diarrhea, diabetes, cancer, HIV/AIDS, liver cirrhosis, alcoholism, anatomic malformations of urinary tract, nephrolithiasis, or urolithiasis were excluded, as were patients with hospital-acquired UTIs. The urine was cultured immediately after collection. Thirty-one UTI patients met the study criteria of having E. faecalis CFU >103/mL isolated from a urine sample in pure culture and were raising poultry in their households.
The urine samples were cultured on Flexicult agar plates (Statens Serum Institut, Copenhagen, Denmark), where E. faecalis grows as small green/blue-green colonies and E. faecium as small green colonies (10). Three colonies were isolated from each UTI patient. All 31 participants were interviewed when urine samples were collected. Personal information recorded included age, sex, and underlying diseases. The following clinical symptoms were recorded: frequent urination, painful urination, cloudy urine, blood in urine, pain in pelvic area, flank pain, pain in low back, and fever. In addition, information about duration of symptoms; previous UTIs; and medical treatment before arrival at the hospital, including type of antimicrobial drug used, was recorded.
Species identification of all 31 presumptive E. faecalis isolates from urine and 83 isolates from poultry were confirmed by species-specific PCR as described by Dutka-Malen et al. (11). Only isolates identified as E. faecalis by PCR were further characterized.
All study participants were informed orally and in writing about the study and provided written consent. The ethics committee at Army Hospital 103 approved the study protocols.
Collection of Cloacal Swabs from Poultry
When a urine sample was positive for E. faecalis, the patient’s household was visited within 1 week, and cloacal swabs were taken from 2–4 chickens in the household. Fecal samples were taken with a sterile cotton swab and immediately placed in Cary-Blair media (Oxoid, Basingstoke, Hampshire, UK) for transportation to the laboratory. Samples were then streaked on Slanetz and Bartley agar medium (Merck, Darmstadt, Germany) the same day and incubated for 24–48 h at 37°C. Subsequently, 2 individual colonies were randomly selected and subcultured on nonselective LB-agar, Lennox plates (Difco, Becton Dickinson, Sparks, MD, USA), which were incubated overnight at 37°C to obtain pure cultures. Colonies were then grown in brain–heart infusion broth (Oxoid) overnight at 37°C and stored for further characterization at –80°C in cryotubes containing 30% glycerol.
MLST and PFGE
To investigate whether isolates of E. faecalis from urine and poultry belonged to identical STs, we characterized isolates from urine and poultry by MLST. Urine isolates were characterized by sequencing of all 7 housekeeping genes used in the MLST scheme: gdh, gyd, pstS, gki, aroE, xpt, and yqil. To confirm that the UTIs were caused by a single strain, 1 additional colony from 9 (29%) of 31 urine samples was characterized by sequencing the gki and yqil genes. Two isolates from each chicken were characterized by sequencing the gki and yqil genes. When sequences of both genes in 2 isolates corresponded to the sequence of the same genes in the urine isolate, which occurred in 11 cases, 1 of the 2 isolates from poultry was randomly selected and further characterized. When gene sequences in only 1 isolate from poultry were identical to the isolate from urine, the isolate was further characterized. Primers and PCR conditions are described on the E. faecalis MLST website (http://efaecalis.mlst.net/). Amplicons were sequenced in both directions by Macrogen (Seoul, South Korea). DNA sequences obtained were assembled using CLC Main Workbench 5.2 software (CLC bio, Aarhus, Denmark) and compared with published alleles, and an ST was assigned to each strain (http://efaecalis.mlst.net/). PFGE was performed as described (12) by using the restriction enzyme smaI (New England BioLabs, Ipswich, MA, USA).
Virulence Genes
The presence and sequence of the following 6 virulence genes were used to further characterize the isolates from urine and poultry: asa1, CylA, efaA, Esp, gelE, and EF0591 (13). After detecting the virulence genes by PCR (13), we sequenced the genes in both directions using Macrogen. DNA sequences were compared, and possible nucleotide differences were calculated by using Smith-Waterman local alignment (EMBOSS) available online from the European Bioinformatics Institute: (www.ebi.ac.uk/).
Antimicrobial Drug Susceptibility Testing
MICs were determined for 16 antimicrobial drugs for comparison analyses by using the Sensititer system (Trek Diagnostics Systems, East Grindstead, UK) according to the manufacturer’s guidelines. These drugs were ampicillin (2–32 μg/mL), avilamycin (4–32 μg/mL), chloramphenicol (2–64 μg/mL), daptomycin (0.25–16 μg/mL), erythromycin (0.5–32 μg/mL), gentamicin (16–1,024 μg/mL), kanamicin (128–2,048 μg/mL), linezolid (0.5–8 μg/mL), moxifloxacin (0.25–8 μg/mL), penicillin (2–32 μg/mL), salinomycin (2–16 μg/mL), streptomycin (64–2,048 μg/mL), quinupristin-dalfopristin (0.25–16 μg/mL), tetracycline (1–32 μg/mL), tigecycline (0.015–2 μg/mL), and vancomycin (1–32 μg/mL).
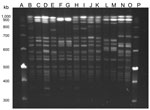
In 7 (23%) of 31 UTI cases, E. faecalis isolated from patient urine and poultry demonstrated identical STs and an indistinguishable (4 pairs) or closely related PFGE pattern (3 pairs, defined as showing <3 fragment difference) (Figure). In addition, antimicrobial drug susceptibility patterns were similar, and only 1 variation was found in the virulence gene profiles (Table 1, Table 2). Five of these 7 patients reportedly had a profession where they worked with poultry. A total of 22 patients who did not share a clone of E. faecalis found in poultry in their household reported working with poultry.
MLST
Sequencing the 7 housekeeping genes in the 31 E. faecalis strains showed the following 14 STs: 4, 16, 17, 93, 116, 136, 141, 314, 410, 411, 412, 413, 415, and 417, with ST16 shown by 16 (51.6%) isolates. Three isolates belonged to ST4, and each of the remaining STs was represented by only 1 isolate. In 7 of 31 households, the same ST was obtained from poultry and urine (Table 1). In 3 households, ST16 was isolated from urine and poultry. In the remaining 4 households, STs 93, 141, 413, and 415 were identified (Table 1). Because each pair of isolates from all selected patients (28%) showed identical gki and yqil gene sequences, we concluded that the UTI cases were associated with 1 E. faecalis strain.
PFGE
We detected 6 PFGE patterns (A1–A6). Of these, 4 pairs from urine and poultry from the same households showed indistinguishable patterns (Table 1; Figure).
Antimicrobial Drug Susceptibility Testing
When we compared isolates from urine and poultry from individual households, we detected similar MICs of each tested antimicrobial drug, showing a 1-dilution factor deviation (Table 2). For several isolates, an MIC could not be established because the MIC fell outside the test intervals. We detected different MICs for 7 antimicrobial drugs when we compared strains 204U and 204P. All isolates were fully susceptible (lowest or second lowest MIC tested) to ampicillin, avilamycin, linezolid, penicillin, salinomycin, tigecycline, and vancomycin (results not shown in Table 2).
Virulence Genes
PCR for the 6 virulence genes showed that the isolates from urine and poultry from an individual household contained identical virulence genes that varied from 1 to 5 genes, except for 1 household in which the isolate from urine (90U) did not contain the asa1 gene (Table 1). When we compared the DNA sequences from the epidemiologically related urine and poultry strains, we found that all 23 sequenced gene pairs showed 100% similarity.
We document isolation of the same clone of E. faecalis in urine and poultry from the same households in which patients had close contact with the poultry. The potential for zoonotic transmission of E. faecalis has been suggested, but to our knowledge, only epidemiologically unrelated isolates have been investigated (3,4,7–9,14).
Most of the isolates in our study belonged to ST16, which has been isolated from animals and humans, including clinical and nonclinical isolates (14). ST93 was isolated from a patient with an ulcer in Poland and from an unknown source in the United States, and ST141 was isolated from chickens in Denmark and from a blood sample of a hospitalized person in Poland (http://efaecalis.mlst.net/).
When we interpreted PFGE patterns for their relatedness using criteria suggested by Tenover et al. (15), we found 4 pairs of E. faecalis strains with indistinguishable band patterns that could be “considered to represent the same strain” (15). From 3 individual households, isolates from urine and poultry showed PFGE patterns with 1 or 2 band differences and thus can be considered closely related (15). These identical or closely related PFGE patterns, together with the supporting findings by MLST and virulence gene profiling, suggest that E. faecalis might be transmitted from poultry to humans, causing UTIs. However, the finding of similar isolates from humans and poultry also could result from sharing a common clone of E. faecalis. ST16 has been reported from various epidemiologically unrelated human and animal sources (14), which could indicate a common clone in humans and animals. Because no data about ST16 in the environment are available, an environmental reservoir cannot be ruled out.
Because 27 of the 31 patients reported having contact with poultry through their work, contact with poultry outside the household environment cannot be excluded as the source of E. faecalis. Epidemiologic risk factor studies are needed to document actual transmission routes.
The variation found in resistance patterns might have resulted from exposure to different antimicrobial drugs, resulting in different selection pressure on E. faecalis in the human and poultry hosts. The 7 patients studied had UTI symptoms for an average of 514 days (range 5 days–10 years), which is unusually long for UTI (Table 1). Although self-medication is well established to be a common practice in Vietnam (16), only 2 of the 7 patients acknowledged use of antimicrobial drugs to treat their UTI symptoms before they participated in the study (data not shown). Over time, patients tend to forget what kind of medication they received. Furthermore, the questionnaire asked only whether antimicrobial drugs were used against UTI, not whether they were used to treat other diseases. In addition, poultry might have been exposed to antimicrobial drugs through growth promoters added in the feedstuff and during therapeutic or preventive treatments, but information about such use was not available.
In most Western countries, contact with poultry occurs mainly through handling and consumption of poultry meat. However, the risk for zoonotic transmission of E. faecalis from poultry meat remains to be investigated. Thus, similar studies and risk factor studies should be conducted in more countries to evaluate the effect on zoonotic transmission of differences in human habits of poultry consumption and contact with poultry. In addition, animals other than pigs and poultry should be investigated as sources of zoonotic E. faecalis transmission. Finally, we cannot exclude the possibility that E. faecalis pathotypes found in poultry might represent transmission from humans, e.g., in this study, from UTI patients. However, poultry as carriers of ST16 has been documented (17), and it seems more likely that humans are exposed to poultry litter than that poultry are exposed to human feces.
We did not investigate the route of E. faecalis transmission, but the route could be colonization of the human intestine and subsequently ascending the urethra as reported for E. coli (18). Further studies are required to explain routes of transmission. The emergence of enterococci as causes of human infections and their resistance to some of the crucial antimicrobial drugs used for human treatment emphasizes the need to elucidate transmission routes and reservoirs for the enterococci and their resistance genes (5,6,19–21).
Dr Poulsen is a PhD student in the Department of Veterinary Disease Biology, Faculty of Health and Medical Sciences, University of Copenhagen. Her primary research interests include zoonotic diseases and transmission routes.
Acknowledgments
We thank all patients for their participation in the study; Flemming Scheutz for his involvement and advice during the development of the protocols; staff at the Army Hospital 103, Ha Dong, Hanoi; and Nina Flint and Gitte Petersen for their excellent technical and laboratory support.
This study was supported by the University of Copenhagen through an ordinary PhD stipend to L.L.P. The Danish International Development Assistance (Danida) provided financial support through the project “Chickens as a possible reservoir for urinary tract infections in humans.”
References
- Murray BE. The life and times of the Enterococcus. Clin Microbiol Rev. 1990;3:46–65.PubMedGoogle Scholar
- Jarvis WR, Martone WJ. Predominant pathogens in hospital infections. J Antimicrob Chemother. 1992;29:19–24.PubMedGoogle Scholar
- Agersø Y, Lester CH, Porsbo LJ, Orsted I, Emborg HD, Olsen KEP, Vancomycin-resistant Enterococcus faecalis isolates from a Danish patient and two healthy human volunteers are possibly related to isolates from imported turkey meat. J Antimicrob Chemother. 2008;62:844–5. DOIPubMedGoogle Scholar
- Donabedian SM, Thal LA, Hershberger E, Perri MB, Chow JW, Bartlett P, Molecular characterization of gentamicin-resistant enterococci in the United States: evidence of spread from animals to humans through food. J Clin Microbiol. 2003;41:1109–13. DOIPubMedGoogle Scholar
- Moellering RC Jr. Emergence of Enterococcus as a significant pathogen. Clin Infect Dis. 1992;14:1173–6. DOIPubMedGoogle Scholar
- Morrison AJ Jr, Wenzel RP. Nosocomial urinary tract infections due to Enterococcus: ten years' experience at a university hospital. Arch Intern Med. 1986;146:1549–51. DOIPubMedGoogle Scholar
- Freitas AR, Coque TM, Novais C, Hammerum AM, Lester CH, Zervos MJ, Human and swine hosts share vancomycin-resistant Enterococcus faecium CC17 and CC5 and Enterococcus faecalis CC2 clonal clusters harboring Tn1546 on indistinguishable plasmids. J Clin Microbiol. 2011;49:925–31. DOIPubMedGoogle Scholar
- Hammerum AM, Lester CH, Heuer OE. Antimicrobial-resistant enterococci in animals and meat: a human health hazard? Foodborne Pathog Dis. 2010;7:1137–46. DOIPubMedGoogle Scholar
- Larsen J, Schonheyder HC, Lester CH, Olsen SS, Porsbo LJ, Garcia-Migura L, Porcine-origin gentamicin-resistant Enterococcus faecalis in humans, Denmark. Emerg Infect Dis. 2010;16:682–4.PubMedGoogle Scholar
- Blom M, Sorensen TL, Espersen F, Frimodt-Moller N. Validation of FLEXICULT SSI-urinary kit for use in the primary health care setting. Scand J Infect Dis. 2002;34:430–5. DOIPubMedGoogle Scholar
- Dutka-Malen S, Evers S, Courvaline P. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J Clin Microbiol. 1995;33:24–7.PubMedGoogle Scholar
- Murray BE, Singh KV, Heath JD, Sharma BR, Weinstock GM. Comparison of genomic DNAs of different enterococcal isolates using restriction endonucleases with infrequent recognition sites. J Clin Microbiol. 1990;28:2059–63.PubMedGoogle Scholar
- Creti R, Imperi M, Bertuccini L, Fabretti F, Orefici G, Di Rosa R, Survey for virulence determinants among Enterococcus faecalis isolated from different sources. J Med Microbiol. 2004;53:13–20. DOIPubMedGoogle Scholar
- Ruiz-Garbajosa P, Bonten MJM, Robinson DA, Top J, Nallapareddy SR, Torres C, Multilocus sequence typing scheme for Enterococcus faecalis reveals hospital-adapted genetic complexes in a background of high rates of recombination. J Clin Microbiol. 2006;44:2220–8. DOIPubMedGoogle Scholar
- Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Interpreting chromosomal dna restriction patterns produced by pulsed-field gel-electrophoresis—criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–9.PubMedGoogle Scholar
- Okumura J, Wakai S. Umenai. Drug utilisation and self-medication in rural communities in Vietnam. Soc Sci Med. 2002;54:1875–86. DOIPubMedGoogle Scholar
- Gregersen RH, Petersen A, Christensen H, Bisgaard M. Multilocus sequence typing of Enterococcus faecalis isolates demonstrating different lesion types in broiler breeders. Avian Pathol. 2010;39:435–40. DOIPubMedGoogle Scholar
- Moreno E, Andreu A, Pigrau C, Kuskowski MA, Johnson JR, Prats G. Relationship between Escherichia coli strains causing acute cystitis in women and the fecal E. coli population of the host. J Clin Microbiol. 2008;46:2529–34. DOIPubMedGoogle Scholar
- Bonten MJ, Willems R, Weinstein RA. Vancomycin-resistant enterococci: why are they here, and where do they come from? Lancet Infect Dis. 2001;1:314–25. DOIPubMedGoogle Scholar
- Hegstad K, Mikalsen T, Coque TM, Werner G, Sundsfjord A. Mobile genetic elements and their contribution to the emergence of antimicrobial resistant Enterococcus faecalis and Enterococcus faecium. Clin Microbiol Infect. 2010;16:541–54. DOIPubMedGoogle Scholar
- Schouten MA, Voss A, Hoogkamp-Korstanje JAA. Antimicrobial susceptibility patterns of enterococci causing infections in Europe. Antimicrob Agents Chemother. 1999;43:2542–6.PubMedGoogle Scholar
Figure
Tables
Cite This ArticleTable of Contents – Volume 18, Number 7—July 2012
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Address for correspondence. Anders Dalsgaard, Department of Veterinary Disease Biology, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, Denmark
Top