Volume 19, Number 6—June 2013
Dispatch
Novel Poxvirus in Big Brown Bats, Northwestern United States
Figure 2
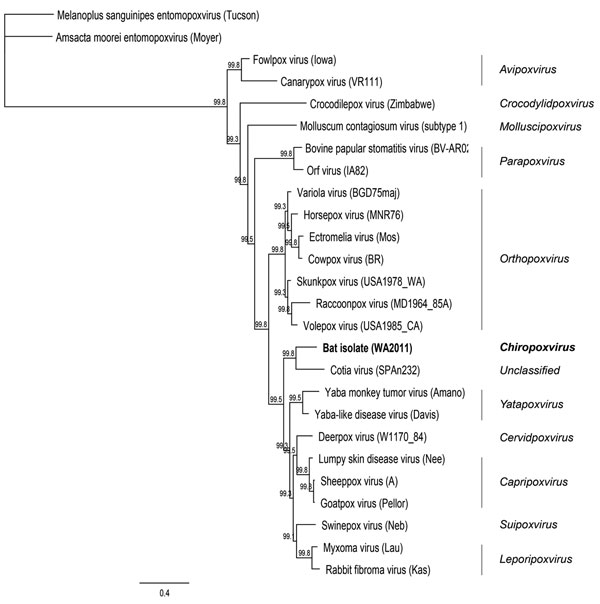
Figure 2. . . Maximum clade credibility tree generated by MrBayes in Geneious version 6.0.5 (www.geneious.com/) using amino acid sequences from 7 open reading frames (final chain length 240,000 at an average SD of split frequencies of zero) for poxviruses. Clade credibility values are indicated at each node.The virus isolated in this study is shown in boldface. Amsacta moorei (red hairy caterpiller) entomopoxvirus (Moyer) was used as the outgroup. Scale bar indicates amino acid substitutions per site.
Page created: April 24, 2013
Page updated: April 24, 2013
Page reviewed: April 24, 2013
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.