Volume 19, Number 9—September 2013
Research
Antigenic and Molecular Characterization of Avian Influenza A(H9N2) Viruses, Bangladesh
Figure 4
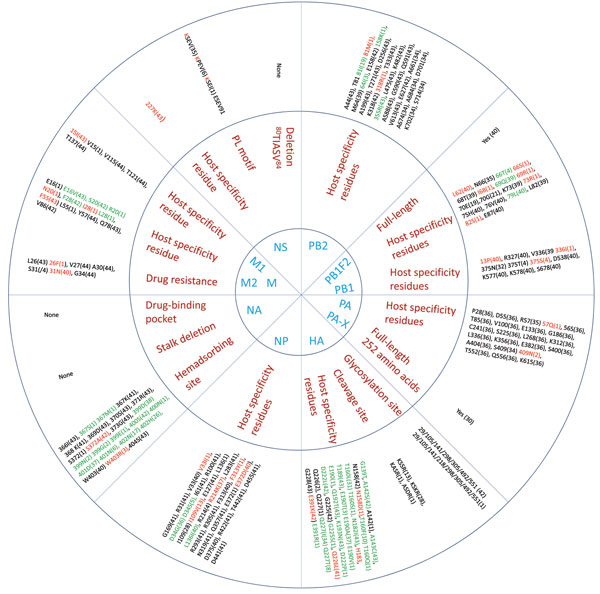
Figure 4. . Host range and pathogenicity determinants in avian influenza (H9N2) viruses isolated from different poultry species in Bangladesh during 2008–2011. Numbers in parentheses indicate number of viruses containing specific amino acid residues of the 44 virus isolates analyzed. Blue indicates the 11 genes that were assessed; red indicates the residues that are critical for influenza pathogenesis, enhanced replication in mammalian hosts, or those that are identical to residues present in human influenza viruses; green indicates unique substitutions in the viruses. HA, hemagglutinin; NA, neuraminidase; M, matrix; NS, nonstructural; NP, nucleoprotein; PA and PB, polymerase genes.
Page created: August 20, 2013
Page updated: August 20, 2013
Page reviewed: August 20, 2013
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.