Volume 20, Number 4—April 2014
Dispatch
Pathology of US Porcine Epidemic Diarrhea Virus Strain PC21A in Gnotobiotic Pigs
Cite This Article
Citation for Media
Abstract
To understand the progression of porcine epidemic diarrhea virus infection, we inoculated gnotobiotic pigs with a newly emerged US strain, PC21A, of the virus. At 24–48 hours postinoculation, the pigs exhibited severe diarrhea and vomiting, fecal shedding, viremia, and severe atrophic enteritis. These findings confirm that strain PC21A is highly enteropathogenic.
A highly contagious coronavirus that causes porcine epidemic diarrhea (PED) was first reported in the United States in May 2013 in Iowa. Since then, the virus—porcine epidemic diarrhea virus (PEDV)—has spread rapidly nationwide (1,2). PEDV (family Coronaviridae, genus Alphacoronavirus) was previously reported only in Europe and Asia. The first US outbreaks caused a high number of deaths among suckling pigs and, as a consequence, substantial economic losses (1,2).
Results of PEDV pathogenesis studies using the prototype European PEDV strain, CV777, were reported in the 1980s (3,4). Strain CV777 infections caused intestinal villous atrophy with substantially reduced ratios of villous height to crypt depth (VH:CD) (3,4). Pathogenic features of CV777 are similar to those observed for Asian PEDV strains that circulated in the 1990s (4–6). To understand the progression of PEDV infection, we studied the pathogenesis of the newly emerged US strain, PC21A.
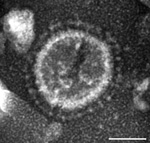
In June 2013, intestinal contents were obtained from a 1-day-old pig with diarrhea on a farm in Ohio, USA. PEDV strain PC21A was detected in the sample by reverse transcription PCR (RT-PCR) selective for the nucleocapsid gene (229–557 nt). The partial nucleocapsid gene sequence of PC21A was identical to that of 2 US PEDV outbreak strains from Colorado, USA: USA/Colorado/2013 (GenBank accession no. KF272920) and 13-019349 (GenBank accession no. KF267450). Only coronavirus-like particles were observed in the fecal sample by electron microscopy (Figure 1). The sample was negative for rotavirus groups A and C and for transmissible gastroenteritis virus/porcine respiratory coronavirus by RT-PCR (7,8).
The sample was bacteriologically sterilized by using 0.22-μm syringe filters and then prepared as inoculum. Near-term gnotobiotic pigs were delivered aseptically by hysterectomy from a specific pathogen–free sow (9). Six 10- to 35-day-old pigs were randomly assigned to a PEDV-infected group (pigs 1–5) or a negative control group (pig 6). Information about inoculation and inocula pig-passage number is described in Table 1. Pigs 1–3 and 5 were inoculated orally and/or intranasally with 6.3–9.0 log10 genomic equivalents (GE) of PEDV strain PC21A; pig 4 was exposed to the virus by indirect contact with inoculated pig 3. For each sample, the quantity of PEDV RNA GE was ≈106 times higher than plaque assay results for a cell-adapted PEDV strain, PC22A. Clinical signs were monitored hourly. Pig 4 was monitored for longer-term clinical signs and virus shedding. Pigs were euthanized for pathologic examination at 3 stages of infection: acute, mid, and later stages (<24 h, 24–48 h, and >48 h, respectively, after onset of clinical signs). The Ohio State University Institutional Animal Care and Use Committee approved all animal-related experimental protocols.
Fecal or rectal swab samples were prepared as described (9). Virus RNA was extracted by using the MagMAX Viral RNA Isolation Kit (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s instructions. Titers of virus shed in feces were determined by TaqMan real-time RT-PCR using the OneStep RT-PCR Kit (QIAGEN, Valencia, CA, USA) as reported (10), with modifications in the forward primer and probe to provide a 100% match to the US strains: forward 5′-CGCAAAGACTGAACCCACTAAC-3′ and probe FAM-TGYYACCAYYACCACGACTCCTGC-BHQ. A standard curve was generated by using the PCR amplicon (PEDN 229/557) of strain PC21A. The detection limit was 10 GE per reaction, corresponding to 4.8 log10 and 3.8 log10 GE/mL of fecal and serum samples, respectively.
Small and large intestine tissues, lung, liver, heart, kidney, spleen, and mesenteric lymph node were examined grossly and histologically. Mean jejunal VH:CD was measured by using PAX-it software (PAXcam, Villa Park, IL, USA) as described (11). The frozen tissues were prepared and tested by immunofluorescence staining, as described (12), for the detection of PEDV antigen, using monoclonal antibody 6C8-1 against the spike protein of PEDV strain DR13 (provided by Daesub Song, Korea Research Institute of Bioscience and Biotechnology, Daejeon, Korea).
Acute, severe watery diarrhea and vomiting developed in all inoculated pigs. Clinical signs developed 24–48 h after inoculation, regardless of the inoculum dose or number of inoculum pig passages (Table 1). Pig 4, which was followed longer, also exhibited dehydration, loss of bodyweight, and lethargy, but it consumed most of the milk that was offered. However, ≈120 h after onset of clinical signs, pig 4 collapsed after showing signs of disorientation and emaciation.
Immune electron microscopy, using a gnotobiotic pig hyperimmune serum to PEDV, showed only PEDV particles in the intestinal contents. For the pig-passaged PC21A strain, RT-PCR/PCR results were negative for transmissible gastroenteritis virus/porcine respiratory coronavirus (7), rotavirus groups A–C (8), caliciviruses (13,14), astroviruses (15), circoviruses, enterovirus, kobuvirus, and bocavirus. For pigs 1 and 2, the detection of fecal virus shedding 24–48 h after inoculation coincided with the onset of clinical signs; for pigs 3 and 4, fecal shedding occurred before the onset of clinical signs (Table 1).
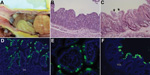
By macroscopic examination, all infected pigs exhibited typical PEDV-like lesions, characterized by thin and transparent intestinal walls (duodenum to colon) and accumulation of large amounts of yellowish fluid in the intestinal lumen (Figure 2, panel A). The stomach was filled with curdled milk, possibly due to reduced intestinal peristalsis. The other internal organs appeared normal. Histologic lesions included acute diffuse, severe atrophic jejunitis (Figure 2, panel B) and mild vacuolation of superficial epithelial cells and subepithelial edema in cecum and colon (Figure 2, panel C). These findings were similar to those in conventional pigs naturally infected with Asian or US strains of PEDV and in caesarean-derived, colostrum-deprived pigs experimentally infected with CV777 (2,3,5,6). The mean jejunal VH:CD of the 5 infected pigs ranged from 1.2 to 3.4, probably depending on the stage of infection (Table 1), and that of the negative control pig was 6.3 (±0.2). VH:CD for pig 4, which was euthanized at a later stage of infection, was 1.5 (±0.2), a ratio indicative of continued cellular necrosis. Neither clinical signs nor lesions developed in the negative control pig during the experiment.
Immunofluorescence-stained cells were observed mainly in the epithelium of atrophied villi of small (duodenum to ileum) and large intestines (Table 2; Figure 2, panels D–F), as reported in other studies (2,3,5). The immunofluorescence was confined to the villous epithelial cells (Figure 2, panels D–F). A few immunofluorescence-stained cells were detected infrequently in the Peyer patches of pig 4. Lung tissues of the infected pigs did not show immunofluorescence staining, indicating that PEDV does not infect lung tissues under the conditions tested. Although PC21A strain replicated in cecum and colon epithelial cells, cellular necrosis and villous atrophy were not evident. Whether PEDV infection of the large intestine contributes to the severity of PED is unclear.
All infected pigs tested at acute or later stages of infection had viral RNA titers of 4.8–7.6 log10 GE/mL in serum samples (Table 1). These titers were similar to those for field samples tested by real-time RT-PCR; 11 (55%) of 20 acute-phase serum samples collected from 13- to 20-week-old pigs with diarrhea from Ohio had viral RNA titers of 4.0–6.3 GE/mL. The early, severe diarrhea and vomiting and the PEDV fecal shedding at high titers may be accompanied by viremia. No infected pigs had detectable viral RNA in serum samples obtained before inoculation, and no negative control pig had detectable viral RNA during the experiment.
In 2013, the first US outbreaks of the rapidly spreading porcine virus, PEDV, caused a high number of pig deaths and substantial economic losses (1,2); however, little was known about progression of the disease. Our data confirm that US PEDV PC21A is highly enteropathogenic and acutely infects the entire intestine, but the jejunum and ileum are the primary sites of infection. PC21A infection causes severe atrophic enteritis accompanied by viremia that leads to severe diarrhea and vomiting.
Dr Jung is a veterinary pathologist at The Ohio State University. His major research interests include diagnostic molecular pathology, pathogenesis, and immune responses to enteric viral infections, using germ-free animal models.
Acknowledgments
We thank James E. Collins and Doug Marthaler for kindly providing US PEDV sequences for primer design and testing for enterovirus, kobuvirus, and bocavirus; J. Hanson, G. Meyers, and R. McComick for assistance with animal care; X. Wang, M. Lee, S.S. Wagner, A. Veeramani, and Chun-Ming Lin for technical assistance; Andrea Kaszas for assistance with electron microscopy; and V. Anastasia for helpful discussion.
Salaries and research support were provided by state and federal funds appropriated to the Ohio Agricultural Research and Development Center, The Ohio State University. This work was supported in part by a grant from the National Pork Board (no. 13-222 to Q.W. and L.J.S.).
References
- Cima G. Fighting a deadly pig disease: industry, veterinarians trying to contain PED virus, new to the US. J Am Vet Med Assoc. 2013;243:469–70 .PubMedGoogle Scholar
- Stevenson GW, Hoang H, Schwartz KJ, Burrough EB, Sun D, Madson D, Emergence of Porcine epidemic diarrhea virus in the United States: clinical signs, lesions, and viral genomic sequences. J Vet Diagn Invest. 2013;25:649–54 . DOIGoogle Scholar
- Debouck P, Pensaert M, Coussement W. The pathogenesis of an enteric infection in pigs, experimentally induced by the coronavirus-like agent, Cv-777. Vet Microbiol. 1981;6:157–65. DOIGoogle Scholar
- Coussement W, Ducatelle R, Debouck P, Hoorens J. Pathology of experimental CV777 coronavirus enteritis in piglets. I. Histological and histochemical study. Vet Pathol. 1982;19:46–56 . DOIGoogle Scholar
- Sueyoshi M, Tsuda T, Yamazaki K, Yoshida K, Nakazawa M, Sato K, An immunohistochemical investigation of porcine epidemic diarrhoea. J Comp Pathol. 1995;113:59–67 . DOIGoogle Scholar
- Kim O, Chae C. In situ hybridization for the detection and localization of porcine epidemic diarrhea virus in the intestinal tissues from naturally infected piglets. Vet Pathol. 2000;37:62–7 . DOIGoogle Scholar
- Kim L, Chang KO, Sestak K, Parwani A, Saif LJ. Development of a reverse transcription–nested polymerase chain reaction assay for differential diagnosis of transmissible gastroenteritis virus and porcine respiratory coronavirus from feces and nasal swabs of infected pigs. J Vet Diagn Invest. 2000;12:385–8 . DOIPubMedGoogle Scholar
- Amimo JO, Vlasova AN, Saif LJ. Detection and genetic diversity of porcine group A rotaviruses in historic (2004) and recent (2011 and 2012) swine fecal samples in Ohio: predominance of the G9P[13] genotype in nursing piglets. J Clin Microbiol. 2013;51:1142–51 . DOIPubMedGoogle Scholar
- Jung K, Wang Q, Kim Y, Scheuer K, Zhang Z, Shen Q, The effects of simvastatin or interferon-alpha on infectivity of human norovirus using a gnotobiotic pig model for the study of antivirals. PLoS ONE. 2012;7:e41619 . DOIPubMedGoogle Scholar
- Kim SH, Kim IJ, Pyo HM, Tark DS, Song JY, Hyun BH. Multiplex real-time RT-PCR for the simultaneous detection and quantification of transmissible gastroenteritis virus and porcine epidemic diarrhea virus. J Virol Methods. 2007;146:172–7 . DOIPubMedGoogle Scholar
- Jung K, Kim J, Ha Y, Choi C, Chae C. The effects of transplacental porcine circovirus type 2 infection on porcine epidemic diarrhoea virus-induced enteritis in preweaning piglets. Vet J. 2006;171:445–50 . DOIPubMedGoogle Scholar
- Jung K, Renukaradhya GJ, Alekseev KP, Fang Y, Tang Y, Saif LJ. Porcine reproductive and respiratory syndrome virus modifies innate immunity and alters disease outcome in pigs subsequently infected with porcine respiratory coronavirus: implications for respiratory viral co-infections. J Gen Virol. 2009;90:2713–23 . DOIPubMedGoogle Scholar
- Sisay Z, Wang Q, Oka T, Saif L. Prevalence and molecular characterization of porcine enteric caliciviruses and first detection of porcine kobuviruses in US swine. Arch Virol. 2013;158:1583–8 . DOIPubMedGoogle Scholar
- Wang Q, Scheuer K, Ahang Z, Gebreyes WA, Molla BZ, Hoet AE, Characterization and prevalence of a new porcine calicivirus in swine, United States. Emerg Infect Dis. 2011;17:1103–6 . DOIPubMedGoogle Scholar
- Chu DK, Poon LL, Guan Y, Peiris JS. Novel astroviruses in insectivorous bats. J Virol. 2008;82:9107–14 . DOIPubMedGoogle Scholar
Figures
Tables
Cite This Article1These authors were co-principal investigators.
Table of Contents – Volume 20, Number 4—April 2014
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Qiuhong Wang, Food Animal Health Research Program, Ohio Agricultural Research and Development Center, Department of Veterinary Preventive Medicine, The Ohio State University, 1680 Madison Ave, Wooster, Ohio 44691, USAQiuhong Wang, Food Animal Health Research Program, Ohio Agricultural Research and Development Center, Department of Veterinary Preventive Medicine, The Ohio State University, 1680 Madison Ave, Wooster, Ohio 44691, USA
Top