Volume 20, Number 4—April 2014
Research
Novel Betacoronavirus in Dromedaries of the Middle East, 2013
Figure 2
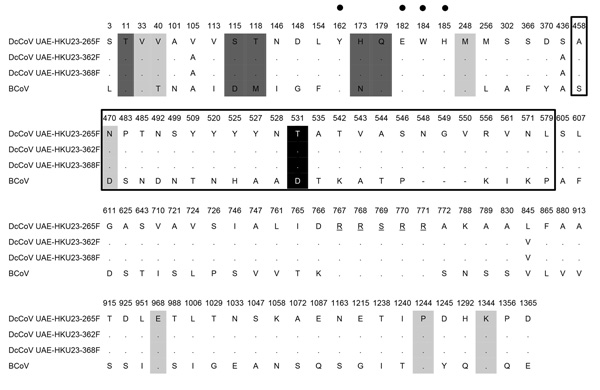
Figure 2. . Amino acid comparison of the spike protein of a novel betacoronavirus, dromedary camel coronavirus (DcCoV) UAE-HKU23, discovered in dromedaries in the Middle East in 2013, with that of bovine coronavirus (BCoV; GenBank accession no. AF391541). Amino acid substitution sites, key amino acids for virulence and receptor binding in BCoV, and cleavage sites are shown. Amino acid positions are given with reference to DcCoV UAE-HKU23. Conserved amino acids, compared with those of DcCoV UAE-HKU23 (strain 265F), are represented by dots. Amino acids of putative cleavage sites are underlined. Amino acids within the S1 hypervariable region of BCoV are marked with open boxes. Amino acid sites central to virulence in BCoV are highlighted in light gray. Amino acid sites shown to affect S1-mediated receptor binding in BCoV are highlighted in dark gray. The 4 critical sugar-binding residues are indicated by black dots. The amino acid site that discriminated between enteric and respiratory BCoV strains is highlighted in black.
1These authors contributed equally to this article.