Volume 20, Number 5—May 2014
Research
Streptococcus mitis Strains Causing Severe Clinical Disease in Cancer Patients
Figure 3
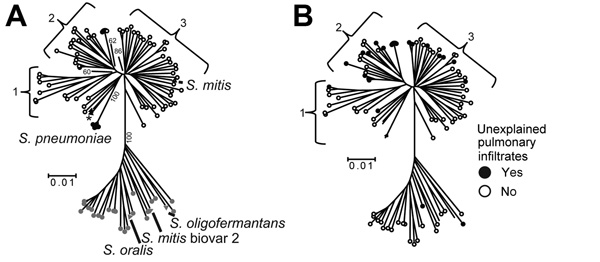
Figure 3. Multilocus sequence analysis (MLSA) and clinical correlates of Streptococcus mitis and Soralis strainsA) For reference purposes, the following are labeled: viridans group streptococci (VGS) strains (SK142 for Smitis, SK23 for Soralis, SK1136 for Soligofermantans), 5 Spneumoniae strains, 2 Spseudopneumoniae strains (SK674 and 103, indicated by an asterisk), and strain SK96 (previously characterized as an Smitis biovar 2 strain)Numbers within the tree refer to bootstrap support values (%).B) For reference purposes, branches of the previously labeled VGS, Spneumoniae, and Spseudopneumoniae strains have been retained; however, for clarity, the branches are not labeledThe presence or absence of unexplained pulmonary infiltrates is indicated as described in the keyBootstrap support values are the same as in panel ANumbers 1–3 indicate Smitis clusters, and scale bars indicate genetic distances.