Volume 20, Number 8—August 2014
Research
Global and Local Persistence of Influenza A(H5N1) Virus
Figure 2
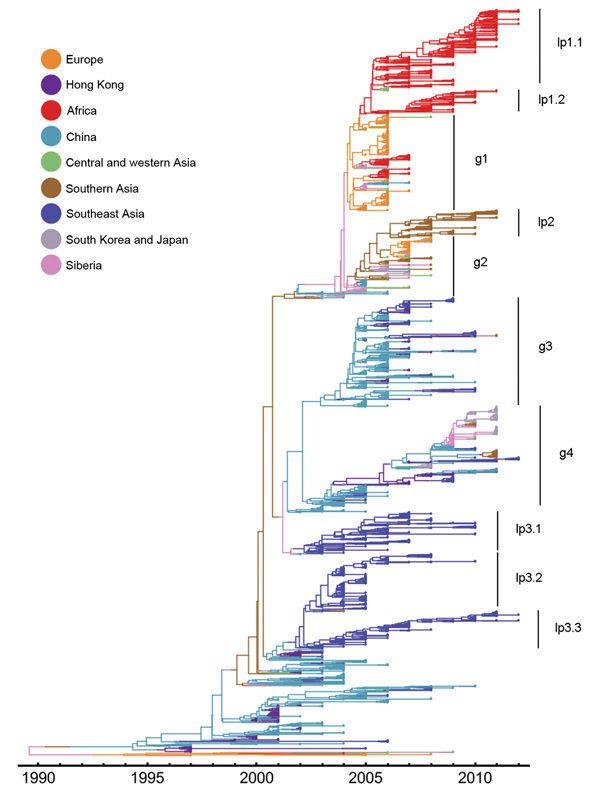
Figure 2. Estimated genealogy of 3,365 highly pathogenic avian influenza A(H5N1) viruses sampled during 1996–2012The maximum a posteriori tree was estimated by using Migrate version 3.3.0 (21,22)Each tip represents a virus sequenceColors indicate the sampling region, either actual (tips) or estimated (branches).
References
- Kilpatrick AM, Chmura AA, Gibbons DW, Fleischer RC, Marra PP, Daszak P. Predicting the global spread of H5N1 avian influenza. Proc Natl Acad Sci U S A. 2006;103:19368–73. DOIPubMedGoogle Scholar
- Gauthier-Clerc M, Lebarbenchon C, Thomas F. Recent expansion of highly pathogenic avian influenza H5N1: a critical review. Ibis. 2007;149:202–14. DOIGoogle Scholar
- Altizer S, Bartel R, Han BA. Animal migration and infectious disease risk. Science. 2011;331:296. DOIPubMedGoogle Scholar
- Xu X. Subbarao, Cox NJ, Guo Y. Genetic characterization of the pathogenic influenza A/goose/Guangdong/1/96 (H5N1) virus: similarity of its hemagglutinin gene to those of H5N1 viruses from the 1997 outbreaks in Hong Kong. Virology. 1999;261:15–9. DOIPubMedGoogle Scholar
- World Health Organization. Cumulative number of confirmed human cases of avian influenza A(H5N1) reported to WHO, 2003–2013. 2013 June 4 [cited 2013 Jun 11]. http://www.who.int/influenza/human_animal_interface/EN_GIP_20130604CumulativeNumberH5N1cases.pdf
- Nelson MI, Simonsen L, Viboud C, Miller MA, Holmes EC. Phylogenetic analysis reveals the global migration of seasonal influenza A viruses. PLoS Pathog. 2007;3:1220–8. DOIPubMedGoogle Scholar
- Rambaut A, Pybus OG, Nelson MI, Viboud C, Taubenberger JK, Holmes EC. The genomic and epidemiological dynamics of human influenza A virus. Nature. 2008;453:615–9. DOIPubMedGoogle Scholar
- Russell CA, Jones TC, Barr IG, Cox NJ, Garten RJ, Gregory V, The global circulation of seasonal influenza A (H3N2) viruses. Science. 2008;320:340–6. DOIPubMedGoogle Scholar
- Bedford T, Cobey S, Beerli P, Pascual M. Global migration dynamics underlie evolution and persistence of human influenza A (H3N2). PLoS Pathog. 2010;6:e1000918. DOIPubMedGoogle Scholar
- Chan J, Holmes A, Rabadan R. Network analysis of global influenza spread. PLOS Comput Biol. 2010;6:e1001005. DOIPubMedGoogle Scholar
- Le MQ, Lam HM, Cuong VD, Lam TT, Halpin RA, Wentworth DE, Migration and persistence of human influenza A viruses, Vietnam, 2001–2008. Emerg Infect Dis. 2013;19:1756–65. DOIPubMedGoogle Scholar
- Bahl J, Nelson MI, Chan KH, Chen R, Vijaykrishna D, Halpin RA, Temporally structured metapopulation dynamics and persistence of influenza A H3N2 virus in humans. Proc Natl Acad Sci U S A. 2011;108:19359–64. DOIPubMedGoogle Scholar
- Wu B, Wang C, Dong G, Guo Y, Nolte DL, Deliberto TJ, New evidence suggests southern China as a common source of multiple clusters of highly pathogenic H5N1 avian influenza virus. J Infect Dis. 2010;202:452–8. DOIPubMedGoogle Scholar
- Wallace RG, Hodac H, Lathrop RH, Fitch WM. A statistical phylogeography of influenza A H5N1. Proc Natl Acad Sci U S A. 2007;104:4473–8. DOIPubMedGoogle Scholar
- Wallace RG, Fitch WM. Influenza A H5N1 immigration is filtered out at some international borders. PLoS ONE 2008;3(2):e1697. PMID: 18301773 DOIGoogle Scholar
- Smith GJ, Fan XH, Wang J, Li KS, Qin K, Zhang JX, Emergence and predominance of an H5N1 influenza variant in China. Proc Natl Acad Sci U S A. 2006;103:16936–41. DOIPubMedGoogle Scholar
- Wang J, Vijaykrishna D, Duan L, Bahl J, Zhang JX, Webster RG, Identification of the progenitors of Indonesian and Vietnamese avian influenza A (H5N1) viruses from southern China. J Virol. 2008;82:3405–14. DOIPubMedGoogle Scholar
- Guan Y, Smith GJ, Webby R, Webster RG. Molecular epidemiology of H5N1 avian influenza. Rev Sci Tech. 2009;28:39–47 .PubMedGoogle Scholar
- Liang L, Xu B, Chen Y, Liu Y, Cao W, Fang L, Combining spatial-temporal and phylogenetic analysis approaches for improved understanding on global H5N1 transmission. PLoS ONE. 2010;5:e13575. DOIPubMedGoogle Scholar
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. DOIPubMedGoogle Scholar
- Beerli P, Felsenstein J. Maximum likelihood estimation of a migration matrix and effective population sizes in n subpopulations by using a coalescent approach. Proc Natl Acad Sci U S A. 2001;98:4563–8. DOIPubMedGoogle Scholar
- Beerli P. Comparison of Bayesian and maximum-likelihood inference of population genetic parameters. Bioinformatics. 2006;22:341–5. DOIPubMedGoogle Scholar
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. DOIPubMedGoogle Scholar
- Hudson RR, Slatkin M, Maddison WP. Estimation of levels of gene flow from DNA sequence data. Genetics. 1992;132:583–9 .PubMedGoogle Scholar
- Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. DOIPubMedGoogle Scholar
- Hasegawa M, Kishino H, Yano T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. 1985;22:160–74. DOIPubMedGoogle Scholar
- Parker J, Rambaut A, Pybus OG. Correlating viral phenotypes with phylogeny: accounting for phylogenetic uncertainty. Infect Genet Evol. 2008;8:239–46. DOIPubMedGoogle Scholar
- Brooks SP, Gelman A. General methods for monitoring convergence of iterative simulations. J Comput Graph Stat. 1998;7:434–55 .DOIGoogle Scholar
- Minin VN, Bloomquist EW, Suchard MA. Smooth skyride through a rough skyline: Bayesian coalescent-based inference of population dynamics. Mol Biol Evol. 2008;25:1459–71. DOIPubMedGoogle Scholar
- Nelson MI, Holmes EC. The evolution of epidemic influenza. Nat Rev Genet. 2007;8:196–205. DOIPubMedGoogle Scholar
- Tang JW, Ngai KLK, Lam WY, Chan PKS. Seasonality of influenza A (H3N2) virus: a Hong Kong perspective (1997–2006). PLoS ONE. 2008;3:e2768. DOIPubMedGoogle Scholar
- Liu J, Xiao H, Lei F, Zhu Q, Qin K, Zhang XW, Highly pathogenic H5N1 influenza virus infection in migratory birds. Science. 2005;309:1206. DOIPubMedGoogle Scholar
- Takekawa JY, Prosser DJ, Newman SH, Muzaffar SB, Hill NJ, Yan B, Victims and vectors: highly pathogenic avian influenza H5N1 and the ecology of wild birds. Avian Biology Research. 2010;3:51–73.
- Si Y, Skidmore AK, Wang T, de Boer WF, Debba P. Spatiotemporal dynamics of global outbreaks match bird migration patterns. Geospat Health. 2009;4:65–78 .PubMedGoogle Scholar
- Prosser DJ, Cui P, Takekawa JY, Tang M, Hou Y, Collins BM, Wild bird migration across the Qinghai-Tibetan Plateau: a potential transmission route for highly pathogenic H5N1. PLoS ONE. 2011;6:e17622. DOIPubMedGoogle Scholar
- Chen H, Li Y, Li Z, Shi J, Shinya K, Deng G, Properties and dissemination of H5N1 viruses isolated during an influenza outbreak in migratory waterfowl in western China. J Virol. 2006;80:5976–83. DOIPubMedGoogle Scholar
- Newman SH, Hill NJ, Spragens KA, Janies D, Voronkin IO, Prosser DJ, Eco-virological approach for assessing the role of wild birds in the spread of avian influenza H5N1 along the Central Asian flyway. PLoS ONE. 2012;7:e30636. DOIPubMedGoogle Scholar
- Guan Y, Webby R, Capua I, Waldenström J. H5N1: how to track a flu virus. Nature. 2012;483:535–6. DOIPubMedGoogle Scholar
Page created: July 18, 2014
Page updated: July 18, 2014
Page reviewed: July 18, 2014
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.