Methicillin-Susceptible, Vancomycin-Resistant Staphylococcus aureus, Brazil
Diana Panesso, Paul Planet, Lorena Diaz, Jean-Emmanuel Hugonnet, Truc T. Tran, Apurva Narechania, Jose M. Munita, Sandra Rincon, Lina P. Carvajal, Jinnethe Reyes, Alejandra Londoño, Hannah Smith, Robert Sebra, Gintaras Deikus, George M. Weinstock, Barbara E. Murray, Flavia Rossi, Michel Arthur, and Cesar A. Arias

Author affiliations: Universidad El Bosque, Bogota, Colombia (D. Panesso, P.J. Planet, L. Diaz, J.M. Munita, S. Rincon, L.P. Carvajal, J. Reyes, G. M. Weinstock, C.A. Arias); University of Texas Medical School, Houston, Texas, USA (D. Panesso, T.T. Tran, J.M. Munita, A. Londoño, B.E. Murray, C.A. Arias); American Museum of Natural History, New York (P.J. Planet, A. Narechania); Columbia University, New York, New York, USA (P.J. Planet, A. Londoño, H. Smith); Centre de Recherche des Cordeliers, Paris, France (J.E. Hugonnet, M. Arthur); Université Pierre et Marie Curie, Paris (J.E. Hugonnet, M. Arthur); Université Paris Descartes, Paris (J.E. Hugonnet, M. Arthur); Clinica Alemana de Santiago, Santiago, Chile (J.M. Munita); Universidad del Desarrollo, Santiago (J.M. Munita); Icahn School of Medicine at Mount Sinai, New York (R. Sebra, G. Deikus); The Jackson Laboratory for Genomic Medicine, Farmington, Connecticut, USA (G.M. Weinstock); Universidade da São Paulo, São Paulo, Brazil (F. Rossi)
Main Article
Figure 2
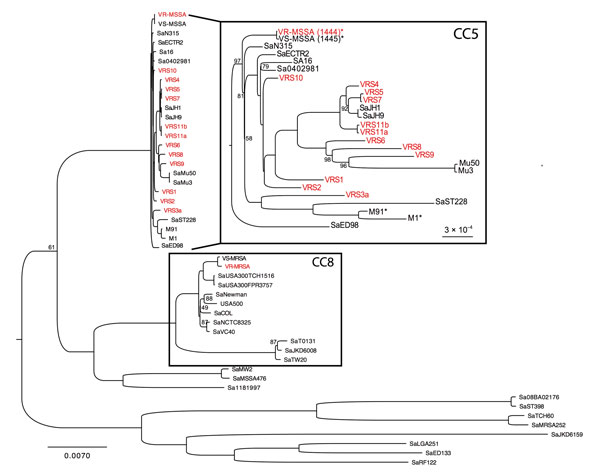
Figure 2. Phylogenetic analyses of Staphylococcus aureus strains, Brazil. Whole-genome phylogenetic tree (dataset = 325,732 single-nucleotide polymorphisms, gamma-based log likelihood − 1909607.06950) of the S. aureus species showing position of vancomycin-resistant, methicillin-susceptible S. aureus (VR-MSSA) and vancomycin-susceptible MSSA (VS-MSSA) isolates sequenced for this study. Vancomycin-resistant S. aureus (VRSA) strains are shown in red. Numbers on branches are bootstrap values based on 1,000 resampling iterations. All branches without numbers had bootstrap values of 100%. Branch lengths are proportional to number of nucleotide substitutions per site (scale bars). Inset labeled CC5 is expanded to emphasize the polyphyly of VRSA strains. *Genomes sequenced for this study. M1 and M91 are members of the Chilean/Cordobes clone that is widespread in Latin America (Technical Appendix). CC, clonal complex.
Main Article
Page created: September 22, 2015
Page updated: September 22, 2015
Page reviewed: September 22, 2015
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
