Volume 21, Number 10—October 2015
Dispatch
Methicillin-Susceptible, Vancomycin-Resistant Staphylococcus aureus, Brazil
Cite This Article
Citation for Media
Abstract
We report characterization of a methicillin-susceptible, vancomycin-resistant bloodstream isolate of Staphylococcus aureus recovered from a patient in Brazil. Emergence of vancomycin resistance in methicillin-susceptible S. aureus would indicate that this resistance trait might be poised to disseminate more rapidly among S. aureus and represents a major public health threat.
Acquisition of high-level vancomycin resistance by Staphylococcus aureus represents a major public health risk because this antimicrobial drug continues to be the first-line and most inexpensive therapy to treat methicillin-resistant S. aureus (MRSA) despite concerns about its clinical efficacy. Recently, we described vancomycin-resistant MRSA (VR-MRSA) recovered from the bloodstream of a patient in Brazil (1). VR-MRSA belongs to sequence type (ST) 8 and is phylogenetically related to the community-associated (CA) MRSA USA300 genetic lineage that has rapidly disseminated in the United States and the northern region of South America (USA300-Latin American variant [USA300-LV]) (1,2). The vanA gene cluster in VR-MRSA was carried by a transferable staphylococcal plasmid (pBRZ01). We characterized a clinical isolate of vancomycin-resistant, methicillin-susceptible S. aureus (VR-MSSA) and document the in vivo transfer of the vanA gene cluster to 2 unrelated S. aureus strains causing bacteremia within the same patient.
On August 28, 2012, a blood culture from a patient in Brazil was reported positive for 2 isolates of MSSA while the patient was receiving daptomycin therapy (Technical Appendix). One MSSA isolate was susceptible to all antimicrobial drugs tested (VS-MSSA). The second isolate (VR-MSSA) had a vancomycin MIC of 256 µg/mL and was also resistant to gentamicin (Table 1). Both isolates were susceptible to daptomycin (MIC 0.5 μg/mL). Thirteen days earlier, 2 MRSA isolates, 1 of which was resistant to vancomycin (VR-MRSA), were recovered from the blood of the same patient (Technical Appendix) (1). The daptomycin MICs for both MRSA strains were also 0.5 μg/mL.
Bacterial strains used in this study (Table 1) were grown in brain–heart infusion broth and agar. Plasmid pBRZ01 was transferred by using filter mating (3) and VR-MSSA and VR-MRSA as donors and VS-MSSA, VS-MRSA, and RN4220RF as recipients (Table 1). Transconjugants were selected on brain heart infusion medium containing vancomycin (32 µg/mL) and fusidic acid (25 µg/mL). Colonies from each mating experiment were subjected to digestion with SmaI and pulsed-field gel electrophoresis to investigate genetic relatedness (1). Plasmids carrying the vanA gene cluster were detected by using S1 nuclease digestion followed by hybridization with a vanA probe (4).
Whole-genome sequencing of VR-MSSA, VS-MSSA, and 2 representatives of the Chilean/Cordobes clone (M1, M91) was performed by using MiSeq PacBio RS II (Illumina, San Diego, CA, USA) to close the VR-MSSA genome (5) (online Technical Appendix). Phylogenetic analysis was performed by using the maximum-likelihood framework within RAxML v7.4.2 (6). For cell wall analysis, extraction and separation of peptidoglycan precursors was performed as described (7).
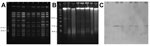
The PFGE patterns of both isolates (VR-MSSA and VS-MSSA) were indistinguishable, and in vitro growth rates were similar (Figure 1, panel A). S1 nuclease analyses indicated that VR-MSSA harbored a plasmid of ≈55 kb, which yielded a positive result when hybridized with a vanA probe (Figure 1, panels B, C) and was similar in size to the previously described vanA-containing plasmid pBRZ01 identified in the same patient (1). pBRZ01 of VR-MSSA was readily transferred to S. aureus RN4220-RF (efficiency = 3 × 10−5/donor). In vitro conjugative transfer of pBRZ01 between MRSA and MSSA strains recovered from the patient’s bloodstream was also readily achieved with efficiencies ranging from 4.3 × 10−7/donor to 2.5 × 10−6/donor. Acquisition of the pBRZ01 by corresponding strains resulted in resistance to vancomycin and gentamicin (Table 1).
Genome sequencing (Technical Appendix) showed that VR-MSSA and VS-MSSA belong to clonal complex (CC) 5 (sequence type ST5) and harbor staphylococcal protein A (Spa) type t002. VS-MSSA and VR-MSSA have the characteristic CC5 genetic traits described by Kos et al. (8). The genome of VR-MSSA has a 2,906,602-bp chromosome and 3 extrachromosomal elements, including a plasmid of 55,713 bp identical to the previously described vanA-carrying pBRZ01 (1), which also harbors aac(6′)-aph(2′′), which confers gentamicin resistance.
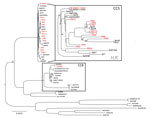
Comparison of the core genomes of VR-MSSA and VS-MSSA showed only 20 single-nucleotide polymorphism differences, which suggested a close genetic relationship and probably representing the same organism that acquired pBRZ01. Phylogenetic analysis (Figure 2) confirmed that VR-MSSA is not a derivative of VR-MRSA (1) (isolated days before from the same patient) and emphasized the relationship of this strain to other vancomycin-resistant S. aureus and MRSA isolates with intermediate susceptibility to vancomycin (VISA).
We analyzed the pool of cytoplasmic peptidoglycan precursors of VR-MSSA grown in the absence or presence of 50 μg/mL of vancomycin for induction of the vanA cluster (Table 2). Tandem mass spectrometry analysis identified 3 nucleotide precursors ending in
Analyses of cell wall muropeptides from VR-MSSA showed 2 modifications of the
In this study, we demonstrated that the vanA-containing pBRZ01 plasmid previously described in MRSA was acquired by an invasive MSSA isolate within the same patient. Our findings also suggest that a vanA-containing plasmid (pBRZ01) was horizontally acquired at least twice during a short period by distinct S. aureus lineages within the same host (MRSA belonging to ST8 and an ST5 MSSA). VR-MSSA belongs to the ST5 lineage of CC5, a major hospital-associated lineage (10). The prevalent hospital-associated lineages circulating in Brazil are ST5 (New York/Japan and Pediatric clones), ST239 (Brazilian clone) and ST1 (USA400 clone) (11), and recent epidemiologic data showed replacement of the endemic Brazilian (ST239) clone by ST5 strains (11–13). Moreover, VR-MSSA is related to ST5 vancomycin-resistant S. aureus strains recovered in the United States (8) and to VISA isolates, including Mu50 and the hetero-VISA strain Mu3, initially recovered in Japan (14). It remains unclear why CC5 strains appear more likely to exhibit vancomycin resistance.
Our biochemical analysis indicates that the vanA gene cluster is fully functional in VR-MSSA, which leads to vancomycin-inducible production of
In summary, we report the in vivo acquisition of high-level vancomycin resistance in a bloodstream MSSA isolate. Of note, vanA-containing pBRZ01 was maintained even after the selective pressure of vancomycin had been removed, raising serious concerns about the possibility of further spread of resistance to this agent. However, no other MSSA strains containing this plasmid have been isolated so far in Brazil.
Dr. Panesso is a postdoctoral researcher at the Laboratory for Antimicrobial Research, University of Texas Medical School at Houston and associate professor of research at the Molecular Genetics and Antimicrobial Resistance Unit, Universidad El Bosque, Bogota, Colombia. Her research interests include the molecular aspects of antimicrobial resistance, with emphasis on gram-positive bacteria.
Acknowledgment
C.A.A. is supported by National Institutes of Health–National Institute of Allergy and Infectious Diseases (NIH-NIAID) grant R01 AI093749, B.E.M. is supported by NIH-NIAID grant R01 AI047923, and P.J.P. is supported by NIH-NIAID grant K08AI101005.
References
- Rossi F, Diaz L, Wollam A, Panesso D, Zhou Y, Rincon S, Transferable vancomycin resistance in a community-associated MRSA lineage. N Engl J Med. 2014;370:1524–31. DOIPubMedGoogle Scholar
- Reyes J, Rincon S, Diaz L, Panesso D, Contreras GA, Zurita J, Dissemination of methicillin-resistant Staphylococcus aureus USA300 sequence type 8 lineage in Latin America. Clin Infect Dis. 2009;49:1861–7. DOIPubMedGoogle Scholar
- Tomita H, Pierson C, Lim SK, Clewell DB, Ike Y. Possible connection between a widely disseminated conjugative gentamicin resistance (pMG1-like) plasmid and the emergence of vancomycin resistance in Enterococcus faecium. J Clin Microbiol. 2002;40:3326–33. DOIPubMedGoogle Scholar
- Arias CA, Panesso D, Singh KV, Rice LB, Murray BE. Cotransfer of antibiotic resistance genes and a hylEfm-containing virulence plasmid in Enterococcus faecium. Antimicrob Agents Chemother. 2009;53:4240–6. DOIPubMedGoogle Scholar
- Benson MA, Ohneck EA, Ryan C, Alonzo F III, Smith H, Narechania A, Evolution of hypervirulence by a MRSA clone through acquisition of a transposable element. Mol Microbiol. 2014;93:664–81. DOIPubMedGoogle Scholar
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–90. DOIPubMedGoogle Scholar
- Bouhss A, Josseaume N, Severin A, Tabei K, Hugonnet JE, Shlaes D, Synthesis of the L-alanyl-L-alanine cross-bridge of Enterococcus faecalis peptidoglycan. J Biol Chem. 2002;277:45935–41. DOIPubMedGoogle Scholar
- Kos VN, Desjardins CA, Griggs A, Cerqueira G, Van Tonder A, Holden MT, Comparative genomics of vancomycin-resistant Staphylococcus aureus strains and their positions within the clade most commonly associated with methicillin-resistant S. aureus hospital-acquired infection in the United States. MBio. 2012;3:e00112–12. DOIPubMedGoogle Scholar
- Arbeloa A, Hugonnet JE, Sentilhes AC, Josseaume N, Dubost L, Monsempes C, Synthesis of mosaic peptidoglycan cross-bridges by hybrid peptidoglycan assembly pathways in gram-positive bacteria. J Biol Chem. 2004;279:41546–56. DOIPubMedGoogle Scholar
- Nübel U, Roumagnac P, Feldkamp M, Song JH, Ko KS, Huang YC, Frequent emergence and limited geographic dispersal of methicillin-resistant Staphylococcus aureus. Proc Natl Acad Sci U S A. 2008;105:14130–5. DOIPubMedGoogle Scholar
- Caboclo RM, Cavalcante FS, Iorio NL, Schuenck RP, Olendzki AN, Felix MJ, Methicillin-resistant Staphylococcus aureus in Rio de Janeiro hospitals: dissemination of the USA400/ST1 and USA800/ST5 SCCmec type IV and USA100/ST5 SCCmec type II lineages in a public institution and polyclonal presence in a private one. Am J Infect Control. 2013;41:e21–6. DOIPubMedGoogle Scholar
- Dabul AN, Kos VN, Gilmore MS, Camargo IL. Draft genome sequence of methicillin-resistant Staphylococcus aureus strain SA16, representative of an endemic clone from a Brazilian hospital. Genome Announc. 2013;1:e00754–13.
- Teixeira MM, Araujo MC, Silva-Carvalho MC, Beltrame CO, Oliveira CC, Figueiredo AM, Emergence of clonal complex 5 (CC5) methicillin-resistant Staphylococcus aureus (MRSA) isolates susceptible to trimethoprim-sulfamethoxazole in a Brazilian hospital. Braz J Med Biol Res. 2012;45:637–43. DOIPubMedGoogle Scholar
- Hiramatsu K, Aritaka N, Hanaki H, Kawasaki S, Hosoda Y, Hori S, Dissemination in Japanese hospitals of strains of Staphylococcus aureus heterogeneously resistant to vancomycin. Lancet. 1997;350:1670–3 . DOIPubMedGoogle Scholar
- Arthur M, Depardieu F, Cabanie L, Reynolds P, Courvalin P. Requirement of the VanY and VanX D,D-peptidases for glycopeptide resistance in enterococci. Mol Microbiol. 1998;30:819–30 . DOIPubMedGoogle Scholar
Figures
Tables
Cite This ArticleTable of Contents – Volume 21, Number 10—October 2015
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Cesar A. Arias, University of Texas Medical School at Houston, 6431 Fannin St, MSB 2.112. Houston, TX 77030, USA
Top