Volume 21, Number 12—December 2015
Research
Zoonotic Leprosy in the Southeastern United States
Figure 2
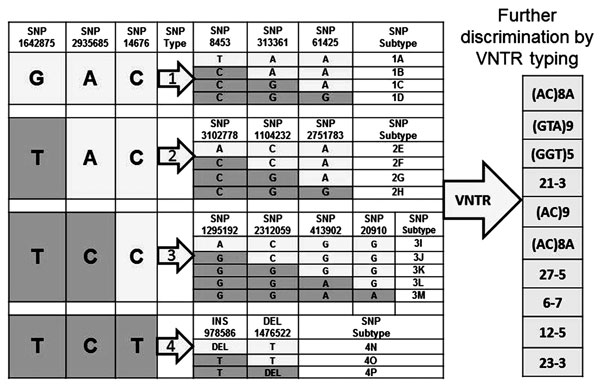
Figure 2. Genotyping scheme for Mycobacterium leprae determined by using single-nucleotide polymorphisms (SNPs) and variable number tandem repeat (VNTR) polymorphisms, southeastern United States. Shading indicates the base that differentiates SNP type and subtype of M. leprae. The algorithm used for strain typing of M. leprae is based on specific SNP location and type and VNTR copy number at the various locations identified along the chromosome. After identification of the major SNP subtype, M. leprae is further discriminated by using allele numbers at 10 VNTR loci. INS, insertion; DEL, deletion.
Page created: October 29, 2015
Page updated: October 29, 2015
Page reviewed: October 29, 2015
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.