Volume 21, Number 7—July 2015
Dispatch
Detection of Circovirus in Foxes with Meningoencephalitis, United Kingdom, 2009–2013
Cite This Article
Citation for Media
Abstract
A fox circovirus was identified in serum samples from foxes with unexplained neurologic signs by using viral metagenomics. Fox circovirus nucleic acid was localized in histological lesions of the cerebrum by in situ hybridization. Viruses from the family Circoviridae may have neurologic tropism more commonly than previously anticipated.
Circoviruses (family Circoviridae) are nonenveloped, single-stranded, circular DNA (≈2 kb) viruses (1). Two genera, Circovirus and Gyrovirus, are recognized, and an additional genus, Cyclovirus, has been proposed (1,2). Circoviruses have an ambisense genome organization with 2 major inversely arranged open reading frames encoding the rolling circle replication initiator protein gene (Rep) and a capsid protein gene (Cap) (1). A conserved stem–loop structure, required for viral replication, is located between the 5′ ends of the 2 main open reading frames. Circoviruses are thought to exhibit host species specificity and have been detected in various species, including birds, pigs, and dogs (1,3,4). These viruses have been associated with a variety of diseases, including respiratory and enteric disease, dermatitis, and reproductive problems (1,3–5). Recently, many small circular DNA genomes have been described from different hosts by using different methods, including high-throughput sequencing (6). Here we describe the identification, characterization, and prevalence of a newly discovered fox circovirus that was present in serum and brain samples from foxes with unexplained meningoencephalitis in the United Kingdom.
During 2009–2013, a total of 31 adult foxes with signs of a neurologic disorder were brought to the RSPCA Norfolk Wildlife Hospital in East Winch, United Kingdom. The foxes exhibited abnormal behavior, lack of fear, reduced alertness, aimless wandering, circling, facial muscle twitching, hind limb paresis, and visual abnormalities. Cases were only detected when free-living foxes became debilitated and were taken to the wildlife rescue center. Once in captivity, diseased foxes had good appetite and generally survived with no substantial disease progression or death, but they showed no evidence of natural recovery. After a few weeks, the foxes were usually euthanized because they did not respond to (nonspecific) medical treatment. All procedures were performed in compliance with relevant laws and institutional guidelines. Following euthanasia, necropsies were performed according to standard procedures. Samples were stored in 10% neutral buffered formalin and embedded in paraffin, and 4 μm–thick sections were stained with hematoxylin and eosin and evaluated for the presence of histologic lesions.
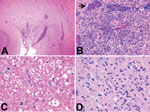
All foxes had similar histologic findings consisting of chronic multifocal or diffuse lymphoplasmacytic meningoencephalitis oriented on the forebrain with a predilection for cortical gray matter (Figure 1; Table 1; Technical Appendix Figure 1). Characteristic histopathologic features were nonspecific perivascular cuffing, rod cell proliferation, spongiosis, neuronal necrosis, moderate to severe gliosis, neuronal satellitosis, and neurophagia. Substantial pathologic changes were restricted to the central nervous system. Histopathologic changes suggested viral, protozoal, microsporidial, immune-mediated, or idiopathic disease. Immunohistochemistry of brain samples was negative for canine distemper virus, canine adenovirus, Borna disease virus, Toxoplasma gondii, and Neospora canium (data not shown). Serologic test results for canine distemper virus, rabies virus, N. canium, and tickborne encephalitis virus were negative, and Ziehl-Neelsen and Giemsa staining results for microsporidia were negative. Minor white matter involvement, the duration of animal survival, and the current absence of documented rabies cases in the United Kingdom eliminated rabies virus as the cause of the neurologic disorder.
Serum samples from 6 of the foxes (VS7100001–6) were available for virus discovery studies. To perform the studies, we used a viral metagenomics approach with the 454 sequence platform (GS Junior; Roche, Basel, Switzerland) as described previously (7–10) (Table 1). More than 22,000 reads were analyzed as described previously (10–12) (Technical Appendix Figure 2). The complete genome sequences of circoviruses from 3 foxes were obtained; the sequences were 99% identical at the nucleotide level (GenBank accession nos. KP260925–7). The fox circovirus genomes had an ambisense organization characteristic of circovirus (Technical Appendix Figure 3). Phylogenetic analysis revealed that the genomes were closely related to those of the recently described canine circoviruses (3,13), displaying ≈92% amino acid identity in the Rep protein and ≈89% nt sequence identity across the entire genome (Technical Appendix Figure 4). On the basis of the suggested criteria demarcating species (1), the fox and canine circoviruses belong to the same species.
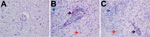
A diagnostic real-time fox circovirus PCR was performed targeting the Rep-coding sequence on 32 serum samples from foxes with and without neurologic signs (Table 1). Viral nucleic acid was extracted by using the MagNA Pure LC Total Nucleic Acid Isolation Kit (Roche, Indianapolis, IN, USA) and amplified by real-time PCR by using primers VS756 (5′-TCCGAGATAGCC GGCGTGGTA-3′), VS757 (5′-CCCGGCCACAGATCAAGTACTTA-3′), and VS758 (5′-FAM-ATCCAACTCCGGAGGAGGAGGA-TAMRA-3′) and the TaqMan Universal Master Mix (Applied Biosystems, Foster City, CA, USA). In addition to samples VS7100001–6, another 14 fox serum samples were positive for fox circovirus, indicating that the virus had infected foxes in multiple counties in the United Kingdom during past years (Table 1; Technical Appendix Figure 5). Clinical data indicated that 77% of circovirus-positive foxes had signs of neurologic disease, compared with only 47% of circovirus-negative foxes (Table 2). Fox circovirus was present in male and female foxes and in adults and juveniles (Table 2). In addition, fox circovirus was detected by real-time PCR in brain samples of 2 of 4 foxes with neurologic disease (VS7100017 and 19; cycle threshold value >35) but not in the brain tissues of 2 foxes without disease. The detection of fox circovirus nucleic acid in the cerebrum of foxes with neurologic disease was confirmed by using the RNAscope 2.0 in situ hybridization kit (Advanced Cell Diagnostics, Hayward, CA, USA) and a Rep gene–specific probe according to the manufacturer’s instructions. Negative controls consisted of circovirus-negative foxes without histopathologic disease. Multifocal fox circovirus RNA signal was detected and associated with the aforementioned histologic lesions in the cerebrum (Figure 2). Specifically, RNA signal was detected in mononuclear cells in perivascular cuffs, inflammatory infiltrates in the neuropil, and neuronal somata in cerebral gray matter of circovirus-positive foxes with neurologic disease. No circovirus signal was found in control foxes with lymphocytic cuffs due to other (known) viral infections or in control foxes without neurologic disease (Figure 2).
Our findings indicate that circoviruses commonly cause systemic infections in wild foxes in the United Kingdom and can be detected in the brains of foxes with neurologic disease. It has been suggested that circoviruses are involved in a plethora of diseases in pigs, dogs, and birds (1,3–5). The canine circovirus may be associated with development of vasculitis in dogs (3), and an overall virus prevalence in serum samples of ≈3% has been reported (3,13). However, we found that the prevalence of fox circovirus in serum samples from foxes with and without neurologic disease was much higher and more comparable to the prevalence of porcine circoviruses among pigs (14). No association of virus infection with vasculitis was apparent. Instead, fox circoviruses may be associated with development of neurologic disease directly or as a contributory complicating cofactor.
Cycloviruses, which belong to a proposed new genus in the family Circoviridae, were recently found in serum and cerebrospinal fluid of humans with paraplegia and acute infections of the central nervous system (11,15), suggesting that viruses from the family Circoviridae may have neurologic tropism more commonly than previously anticipated. However, a causal link between circovirus infection and disease in humans and animals remains to be proven. Because the prevalence of circoviruses in foxes was relatively high and closely related circovirus species seem pathogenic for both dogs and foxes, additional surveillance is warranted to clarify the epidemiology and pathogenicity of circoviruses in foxes.
S.L.S. is employed part time by Viroclinics Biosciences BV.
Mr. Bexton is the senior veterinarian at the RSPCA Norfolk Wildlife Hospital, East Winch, United Kingdom. His main research interests are the epidemiology and pathology of wildlife diseases including novel pathogens in free-living animals.
Acknowledgment
This work was partially funded by the European Commission’s COMPARE H2020 project under grant agreement No 643476, the Virgo Consortium, and ZonMW TOP project 91213058.
References
- Todd D, Bendinelli M, Biagini P, Hino S, Mankertz A, Mishiro S, Circoviridae. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editors. Virus taxonomy: eighth report of the International Committee on Taxonomy of Viruses. San Diego: Elsevier Academic Press; 2005. p. 327–34.
- Li L, Kapoor A, Slikas B, Bamidele OS, Wang C, Shaukat S, Multiple diverse circoviruses infect farm animals and are commonly found in human and chimpanzee feces. J Virol. 2010;84:1674–82. DOIPubMedGoogle Scholar
- Li L, McGraw S, Zhu K, Leutenegger CM, Marks SL, Kubiski S, Circovirus in tissues of dogs with vasculitis and hemorrhage. Emerg Infect Dis. 2013;19:534–41. DOIPubMedGoogle Scholar
- Todd D. Avian circovirus diseases: lessons for the study of PMWS. Vet Microbiol. 2004;98:169–74. DOIPubMedGoogle Scholar
- Opriessnig T, Halbur PG. Concurrent infections are important for expression of porcine circovirus associated disease. Virus Res. 2012;164:20–32. DOIPubMedGoogle Scholar
- Delwart E, Li L. Rapidly expanding genetic diversity and host range of the Circoviridae viral family and other Rep encoding small circular ssDNA genomes. Virus Res. 2012;164:114–21. DOIPubMedGoogle Scholar
- Bodewes R, van der Giessen J, Haagmans BL, Osterhaus AD, Smits SL. Identification of multiple novel viruses, including a parvovirus and a hepevirus, in feces of red foxes. J Virol. 2013;87:7758–64. DOIPubMedGoogle Scholar
- van den Brand JM, van Leeuwen M, Schapendonk CM, Simon JH, Haagmans BL, Osterhaus AD, Metagenomic analysis of the viral flora of pine marten and European badger feces. J Virol. 2012;86:2360–5. DOIPubMedGoogle Scholar
- van Leeuwen M, Williams MM, Koraka P, Simon JH, Smits SL, Osterhaus AD. Human picobirnaviruses identified by molecular screening of diarrhea samples. J Clin Microbiol. 2010;48:1787–94. DOIPubMedGoogle Scholar
- Schürch AC, Schipper D, Bijl MA, Dau J, Beckmen KB, Schapendonk CM, Metagenomic survey for viruses in Western Arctic caribou, Alaska, through iterative assembly of taxonomic units. PLoS ONE. 2014;9:e105227.DOIPubMedGoogle Scholar
- Smits SL, Zijlstra EE, van Hellemond JJ, Schapendonk CM, Bodewes R, Schürch AC, Novel cyclovirus in human cerebrospinal fluid, Malawi, 2010–2011. Emerg Infect Dis. 2013;19:1511-3.DOIPubMedGoogle Scholar
- Prachayangprecha S, Schapendonk CM, Koopmans MP, Osterhaus AD, Schürch AC, Pas SD, Exploring the potential of next-generation sequencing in detection of respiratory viruses. J Clin Microbiol. 2014;52:3722–30. DOIPubMedGoogle Scholar
- Kapoor A, Dubovi EJ, Henriquez-Rivera JA, Lipkin WI. Complete genome sequence of the first canine circovirus. J Virol. 2012;86:7018. DOIPubMedGoogle Scholar
- Rose N, Opriessnig T, Grasland B, Jestin A. Epidemiology and transmission of porcine circovirus type 2 (PCV2). Virus Res. 2012;164:78–89. DOIPubMedGoogle Scholar
- Tan LV, van Doorn HR, Nghia HD, Chau TT, Tu LT, de Vries M, Identification of a new cyclovirus in cerebrospinal fluid of patients with acute central nervous system infections. MBio. 2013;4:e00231–13. DOIPubMedGoogle Scholar
Figures
Tables
Cite This ArticleTable of Contents – Volume 21, Number 7—July 2015
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Saskia L. Smits, Department of Viroscience, Erasmus Medical Center, PO Box 2040, 3000 CA, Rotterdam, the Netherlands
Top