Volume 21, Number 8—August 2015
Research
Influenza A Viruses of Human Origin in Swine, Brazil
Figure 2
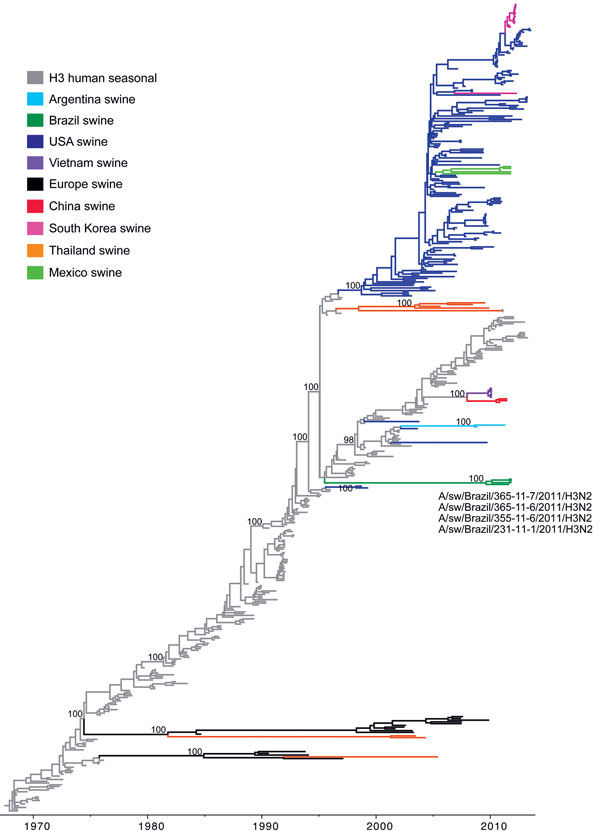
Figure 2. Phylogenetic relationships between human and swine influenza H3 segments. Time-scaled Bayesian maximum clade credibility (MCC) tree inferred for the hemagglutinin (H3) sequences of 463 viruses, including 4 viruses sequenced for this study from swine in Brazil, A/swine/Brazil/365-11-7/2011(H3N2), A/swine/Brazil/231-11-1/2011(H3N2), A/swine/Brazil/355-11-6/2011(H3N2), and A/swine/Brazil/365-11-6/2011(H3N2); 251 human seasonal H3 viruses collected globally during 1968–2013; and 208 closely related swine viruses collected globally that have been studied previously (35). Gray indicates branches of human seasonal influenza A(H3N2) virus origin. Branches associated with viruses from swine are shaded by country/area of origin: light blue, Argentina; dark green, Brazil; dark blue, United States; purple, Vietnam; black, Europe; red, China; pink, Korea; orange, Thailand; light green, Mexico. Posterior probabilities >0.9 are included for key nodes.
References
- Garten RJ, Davis CT, Russell CA, Shu B, Lindstrom S, Balish A, Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science. 2009;325:197–201. DOIPubMedGoogle Scholar
- Chowell G, Echevarría-Zuno S, Viboud C, Simonsen L, Tamerius J, Miller MA, Characterizing the epidemiology of the 2009 influenza A/H1N1 pandemic in Mexico. PLoS Med. 2011;8:e1000436. DOIPubMedGoogle Scholar
- Song MS, Lee JH, Pascua PNQ, Baek YH, Kwon H, Park KJ, Evidence of human-to-swine transmission of the pandemic (H1N1) 2009 influenza virus in South Korea. J Clin Microbiol. 2010;48:3204–11. DOIPubMedGoogle Scholar
- Hofshagen M, Gjerset B, Er C. Pandemic influenza A(H1N1)v: human to pig transmission in Norway. Euro Surveill. 2009;14:1–3 .PubMedGoogle Scholar
- Howden KJ, Brockhoff EJ, Caya FD, McLeod LJ, Lavoie M, Ing JD. An investigation into human pandemic influenza virus (H1N1) 2009 on an Alberta swine farm. Can Vet J. 2009;50:1153–61 .PubMedGoogle Scholar
- Pasma T, Joseph T. Pandemic (H1N1) 2009 infection in swine herds, Manitoba, Canada. Emerg Infect Dis. 2010;16:706–8. DOIPubMedGoogle Scholar
- Trevennec K, Leger L, Lyazrhi F, Baudon E, Cheung CY, Roger F, Transmission of pandemic influenza H1N1 (2009) in Vietnamese swine in 2009–2010. Influenza Other Respir Viruses. 2012;6:348–57.
- Kim SH, Moon OK, Lee KK, Song YK, Yeo CI, Bae CW, Outbreak of pandemic influenza (H1N1) 2009 in pigs in Korea. Vet Rec. 2011;169:155. DOIPubMedGoogle Scholar
- Vijaykrishna D, Poon LLM, Zhu HC, Ma SK, Li OTW, Cheung CL, Reassortment of pandemic H1N1/2009 influenza A virus in swine. Science. 2010;328:1529. DOIPubMedGoogle Scholar
- Starick E, Lange E, Fereidouni S, Bunzenthal C, Höveler R, Kuczka A, Reassorted pandemic (H1N1) 2009 influenza A virus discovered from pigs in Germany. J Gen Virol. 2011;92:1184–8 . DOIPubMedGoogle Scholar
- Ducatez MF, Hause B, Stigger-Rosser E, Darnell D, Corzo C, Juleen K, Multiple reassortment between pandemic (H1N1) 2009 and endemic influenza viruses in pigs, United States. Emerg Infect Dis. 2011;17:1624–9.PubMedGoogle Scholar
- Lam TTY, Zhu H, Wang J, Smith DK, Holmes EC, Webster RG, Reassortment events among swine influenza A viruses in China: implications for the origin of the 2009 influenza pandemic. J Virol. 2011;85:10279–85. DOIPubMedGoogle Scholar
- Liang H, Lam TTY, Fan X, Chen X, Zeng Y, Zhou J, Expansion of genotypic diversity and establishment of 2009 H1N1 pandemic-origin internal genes in pigs in China. J Virol. 2014;88:10864–74. DOIPubMedGoogle Scholar
- Holyoake PK, Kirkland PD, Davis RJ, Arzey KE, Watson J, Lunt RA, The first identified case of pandemic H1N1 influenza in pigs in Australia. Aust Vet J. 2011;89:427–31. DOIPubMedGoogle Scholar
- Nokireki T, Laine T, London L, Ikonen N, Huovilainen A. The first detection of influenza in the Finnish pig population: a retrospective study. Acta Vet Scand. 2013;55:69. DOIPubMedGoogle Scholar
- Njabo KY, Fuller TL, Chasar A, Pollinger JP, Cattoli G, Terregino C, Pandemic A/H1N1/2009 influenza virus in swine, Cameroon, 2010. Vet Microbiol. 2012;156:189–92. DOIPubMedGoogle Scholar
- Pereda A, Cappuccio J, Quiroga M, Baumeister E, Insarralde L, Ibar M, Pandemic (H1N1) 2009 outbreak on pig farm, Argentina. Emerg Infect Dis. 2010;16:304–7. DOIPubMedGoogle Scholar
- Rajão DS, Costa AT, Brasil BS, Del Puerto HL, Oliveira FG, Alves F, Genetic characterization of influenza virus circulating in Brazilian pigs during 2009 and 2010 reveals a high prevalence of the pandemic H1N1 subtype. Influenza Other Respir Viruses. 2013;7:783–90.
- Schaefer R, Zanella JRC, Brentano L, Vincent AL, Ritterbusch GA, Silveira S, Isolation and characterization of a pandemic H1N1 influenza virus in pigs in Brazil. Pesqui Vet Bras. 2011;31:761–7. DOIGoogle Scholar
- Ramirez-Nieto GC, Rojas CAD, Alfonso VJ, Correa JJ, Galvis JD. First isolation and identification of H1N1 swine influenza viruses in Colombian pig farms. Health (Irvine Calif). 2012;04:983–90.
- Escalera-Zamudio M, Cobián-Güemes G, de los Dolores Soto-del Río M, Isa P, Sánchez-Betancourt I, Parissi-Crivelli A, Characterization of an influenza A virus in Mexican swine that is related to the A/H1N1/2009 pandemic clade. Virology. 2012;433:176–82. DOIPubMedGoogle Scholar
- Cappuccio JA, Pena L, Dibárbora M, Rimondi A, Piñeyro P, Insarralde L, Outbreak of swine influenza in Argentina reveals a non-contemporary human H3N2 virus highly transmissible among pigs. J Gen Virol. 2011;92:2871–8. DOIPubMedGoogle Scholar
- Caron LF, Joineau MEG, Santin E, Richartz RRTB, Patricio MAC, Soccol VT. Seroprevalence of H3N2 influenza A virus in pigs from Paraná (South Brazil): interference of the animal management and climatic conditions. Virus Rev Res. 2010;15:63–73. DOIGoogle Scholar
- Cunha RG, Vinha VR, Passos WD. Isolation of a strain of myxovirus influenzae-A suis from swine slaughtered in Rio de Janeiro. Rev Bras Biol. 1978;38:13–7 .PubMedGoogle Scholar
- Rajão DS, Alves F, Del Puerto HL, Braz GF, Oliveira FG, Ciacci-Zanella JR, Serological evidence of swine influenza in Brazil. Influenza Other Respir Viruses. 2013;7:109–12.
- Biondo N, Schaefer R, Gava D, Cantão ME, Silveira S, Mores MAZ, Genomic analysis of influenza A virus from captive wild boars in Brazil reveals a human-like H1N2 influenza virus. Vet Microbiol. 2014;168:34–40. DOIPubMedGoogle Scholar
- Schaefer R, Rech RR, Gava D, Cantão ME, Silva MC, Silveira S. CZJ. A human-like H1N2 influenza virus detected during an outbreak of acute respiratory disease in swine in Brazil. Arch Virol. 2015;160:29–38. DOIPubMedGoogle Scholar
- Lorusso A, Faaberg KS, Killian ML, Koster L, Vincent AL. One-step real-time RT-PCR for pandemic influenza A virus (H1N1) 2009 matrix gene detection in swine samples. J Virol Methods. 2010;164:83–7. DOIPubMedGoogle Scholar
- Zhang J, Harmon KM. RNA extraction from swine samples and detection of influenza A virus in swine by real-time RT-PCR. Methods Mol Biol. 2014;1161:277–93.PubMedGoogle Scholar
- Zhang J, Gauger PC. Isolation of swine influenza virus in cell cultures and embryonated chicken eggs. Methods Mol Biol. 2014;1161:265–76.PubMedGoogle Scholar
- Chan C-H, Lin K-L, Chan Y, Wang Y-L, Chi Y-T, Tu H-L, Amplification of the entire genome of influenza A virus H1N1 and H3N2 subtypes by reverse-transcription polymerase chain reaction. J Virol Methods. 2006;136:38–43. DOIPubMedGoogle Scholar
- Hoffmann E, Stech J, Guan Y, Webster RG, Perez DR. Universal primer set for the full-length amplification of all influenza A viruses. Arch Virol. 2001;146:2275–89. DOIPubMedGoogle Scholar
- Zhou B, Donnelly ME, Scholes DT, St George K, Hatta M, Kawaoka Y, Single-reaction genomic amplification accelerates sequencing and vaccine production for classical and Swine origin human influenza a viruses. J Virol. 2009;83:10309–13. DOIPubMedGoogle Scholar
- Bao Y, Bolotov P, Dernovoy D, Kiryutin B, Zaslavsky L, Tatusova T, The influenza virus resource at the National Center for Biotechnology Information. J Virol. 2008;82:596–601. DOIPubMedGoogle Scholar
- Nelson MI, Wentworth DE, Culhane MR, Vincent AL, Viboud C, LaPointe MP, Introductions and evolution of human-origin seasonal influenza A viruses in multinational swine populations. J Virol. 2014;88:10110–9. DOIPubMedGoogle Scholar
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. DOIPubMedGoogle Scholar
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–90. DOIPubMedGoogle Scholar
- Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29:1969–73. DOIPubMedGoogle Scholar
- Suchard MA, Rambaut A. Many-core algorithms for statistical phylogenetics. Bioinformatics. 2009;25:1370–6. DOIPubMedGoogle Scholar
- Nelson MI, Gramer MR, Vincent AL, Holmes EC. Global transmission of influenza viruses from humans to swine. J Gen Virol. 2012;93:2195–203. DOIPubMedGoogle Scholar
1These authors contributed equally to this article.