Volume 22, Number 1—January 2016
Dispatch
Variations in Spike Glycoprotein Gene of MERS-CoV, South Korea, 2015
Figure 2
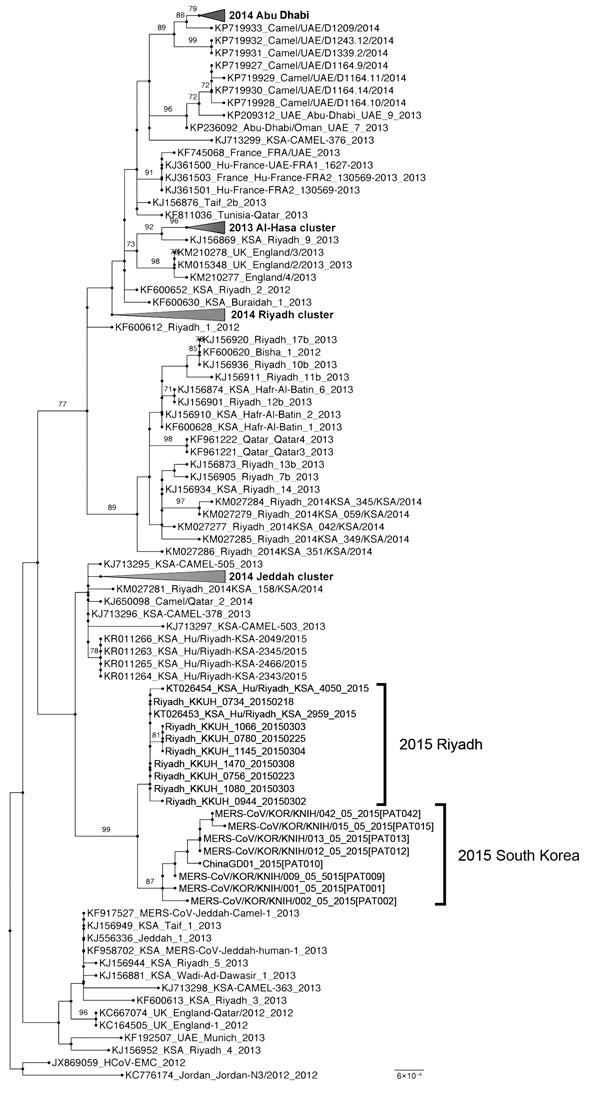
Figure 2. Molecular phylogenetic tree and coding region variants for spike glycoprotein genes of Middle East respiratory syndrome coronavirus (MERS-CoV) isolates from South Korea, May 2015, and reference MERS-CoV sequences. Phylogenetic analysis of 139 spike glycoprotein gene sequences was performed by using RAxML software (10). Tree was visualized with FigTree v.1.4 (http://tree.bio.ed.ac.uk/software/figtree). Taxonomic positions of circulating strains from the outbreak in South Korea and Riyadh are indicated. Compressed major clades of MERS-CoV are indicated in bold. Bootstrap values (>70%) on nodes are shown as percentages on the basis of 1,000 replicates. Scale bar indicates nucleotide substitutions per site.
References
- Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–20 and.PubMedGoogle Scholar
- Raj VS, Mou H, Smits SL, Dekkers DH, Muller MA, Dijkman R, Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–4 and.PubMedGoogle Scholar
- Wang N, Shi X, Jiang L, Zhang S, Wang D, Tong P, Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Res. 2013;23:986–93 and.PubMedGoogle Scholar
- Song F, Fux R, Provacia LB, Volz A, Eickmann M, Becker S, Middle East respiratory syndrome coronavirus spike protein delivered by modified vaccinia virus Ankara efficiently induces virus-neutralizing antibodies. J Virol. 2013;87:11950–4 and.PubMedGoogle Scholar
- World Health Organization WHO). Laboratory testing for Middle East respiratory syndrome coronavirus. Interim guidance (revised). WHO/MERS/LAB/15.1. Geneva: The Organization; 2015 [cited 2015 Oct 15]. http://apps.who.int/iris/bitstream/10665/176982/1/WHO_MERS_LAB_15.1_eng.pdf
- World Health Organization Regional Office for the Western Pacific (WHO/Western Pacific Region). Middle East respiratory syndrome coronavirus (MERS-CoV)—Republic of Korea. Manila: WHO/Western Pacific Region; May 30, 2015 [cited 2015 Oct 5]. http://www.who.int/csr/don/30-may-2015-mers-korea/en/
- Kim YJ, Cho YJ, Kim DW, Yang JS, Kim H, Park S, Complete genome sequence of Middle East respiratory syndrome coronavirus KOR/KNIH/002_05_2015, isolated in South Korea. Genome Announc. 2015;3:e00787–15.PubMedGoogle Scholar
- Somily A, Barry M, Al Subaie SS. BinSaeed AA, Alzamil FA, Zaher W, et al. 2015. Evolution patterns of the Middle East respiratory syndrome coronavirus (MERS-CoV) obtained from MERS patients in early 2015 [cited 2015 Oct 5]. http://virological.org/uploads/default/original/1X/40ca96d0d6ceb95bacbda00b974b50170d412d9b.pdf
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7 and.
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–3 and.
- Mohapatra JK, Pandey LK, Rai DK, Das B, Rodriguez LL, Rout M, Cell culture adaptation mutations in foot-and-mouth disease virus serotype A capsid proteins: implications for receptor interactions. J Gen Virol. 2015;96:553–64 and.
- Cotten M, Watson SJ, Zumla AI, Makhdoom HQ, Palser AL, Ong SH, Spread, circulation, and evolution of the Middle East respiratory syndrome coronavirus. MBio. 2014;5:e01062–13 and.
- Barry MA, Al Subaie SS, Somily AM. BinSaeed AA, Alzamil FA, Al-Jahdali IA, et al. Recent evolution patterns of Middle East respiratory syndrome coronavirus (MERS-CoV), 2015 [cited 2015 Oct 15]. http://virological.org/uploads/default/78/4a6e0a6e90d92867.pdf
1These authors contributed equally to this article.