Volume 23, Number 10—October 2017
Research
Investigation of Outbreaks of Salmonella enterica Serovar Typhimurium and Its Monophasic Variants Using Whole-Genome Sequencing, Denmark
Figure 2
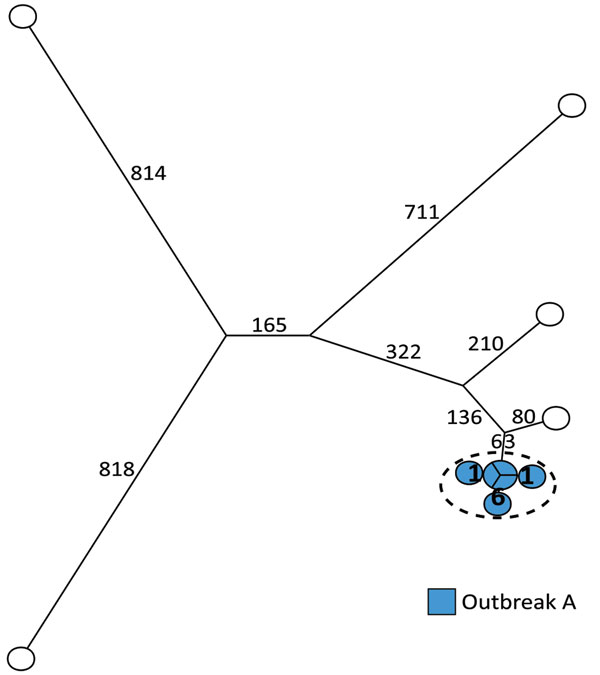
Figure 2. Maximum-parsimony tree of 11 human isolates of Salmonella enterica serovar Typhimurium ST36 based on core-genome SNP analysis with an internal de novo assembled ST36 genome as the reference genome in an outbreak investigation of Salmonella Typhimurium and its monophasic variants, Denmark. Branches are labeled with number of SNP differences. One outbreak (outbreak A) was included. Isolates highlighted in blue belong to outbreak A as previously defined by MLVA; isolates inside the dotted circle are outbreak isolates as defined by the SNP analysis. MLVA, multilocus variable-number tandem-repeat analysis; SNP, single-nucleotide polymorphism; ST, sequence type.
Page created: September 18, 2017
Page updated: September 18, 2017
Page reviewed: September 18, 2017
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.