Volume 23, Number 4—April 2017
Research
Three Divergent Subpopulations of the Malaria Parasite Plasmodium knowlesi
Figure 1
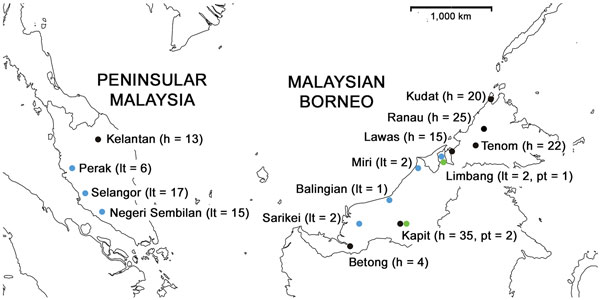
Figure 1. Geographic distribution of DNA samples of Plasmodium knowlesi infections derived from 134 humans and 48 macaques across Malaysia. h, human samples; lt, long-tailed macaque samples; pt, pig-tailed macaque samples.
Page created: March 17, 2017
Page updated: March 17, 2017
Page reviewed: March 17, 2017
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.