Volume 24, Number 1—January 2018
Research
Emergence of Vaccine-Derived Polioviruses during Ebola Virus Disease Outbreak, Guinea, 2014–2015
Figure 2
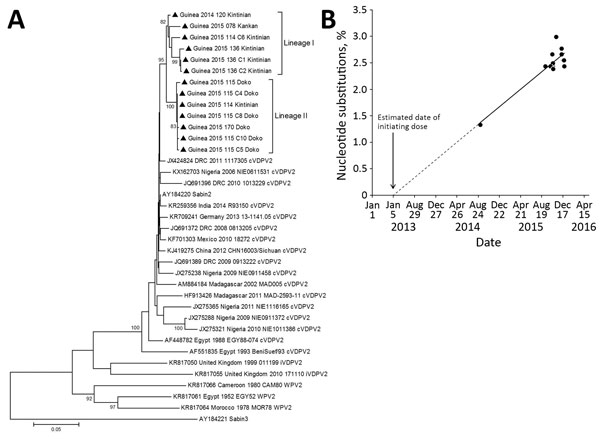
Figure 2. VP1 sequence analysis of cVDPV2 isolated from case-patients (n = 6) and contacts (n = 7) in Guinea, 2014–2015. A) Phylogenetic tree inferred with the complete VP1 region nucleotide sequences (903 bp). Our data were compared with a representative global set of 23 isolates representing type 2 VDPV strains from immunodeficient persons, Sabin-2 strain, cVDPV2s, and wild-type polioviruses identified by GenBank search. The Sabin-3 poliovirus sequence was introduced as an outgroup. The neighbor-joining tree was constructed by using MEGA 5.0 (http://www.megasoftware.net/). Distances were computed by using the Kimura 2-parameter model after excluding positions containing gaps and missing data from the alignments. The robustness of the nodes was tested by 1,000 bootstrap replications. Bootstrap support values >80 are shown in nodes. Triangles indicate the strains from this study. Study strains are indicated by the country, year of isolation, laboratory code, and subprefecture from which isolated. GenBank accession numbers for published sequences are shown in the tree. B) Estimation of the date of initial vaccination with Sabin-2, showing the proportion of VP1 nucleotide changes in the 13 cVDPVs isolates from Guinea with respect to the Sabin-2 reference vaccine strain (AY184220). The data were adjusted to a linear function for the accumulation of nucleotide substitutions (y = 0.0028*x + 1.372; R2 = 0.80). Date of initial OPV infection was calculated by extrapolating the line for the evolution rate of nucleotide substitutions backward to 0% substitutions. Scale bar represents nucleotide substitutions per site. cVDPV2, type-2 circulating vaccine-derived poliovirus; OPV, oral polio vaccine; VP, viral capsid protein.
1These authors contributed equally to this article.