Volume 24, Number 1—January 2018
Research
Emergence of Vaccine-Derived Polioviruses during Ebola Virus Disease Outbreak, Guinea, 2014–2015
Figure 3
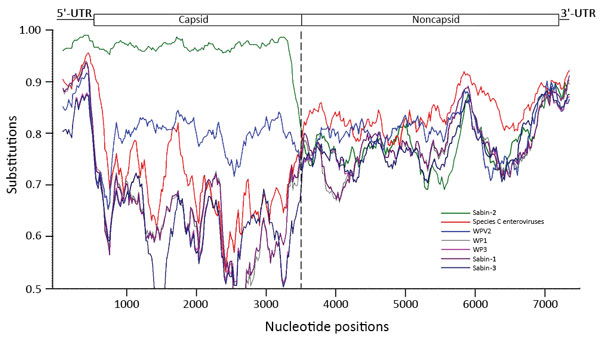
Figure 3. Whole-genome sequence analysis, showing similarity between cVDPV2 isolate 2015-114-C6 (query), 21 prototypes of human enterovirus species C, 3 WPVs, and 3 Sabin poliovirus strains. Approximate nucleotide positions in the poliovirus genome are indicated. The enterovirus genetic map is shown at top. Analyses were calculated by using SimPlot version 3.5 (https://sray.med.som.jhmi.edu/SCRoftware/simplot/). Similarity was calculated in each window of 300 bp by using the Kimura 2-parameter method with a transition:transversion ratio of 2. The window was advanced along the genome alignment in 20-bp increments with 1,000 resamplings. GenBank accession numbers for viruses tested: poliovirus type 1 (V01149), poliovirus type 2 (AY238473 and M12197), poliovirus type 3 (K01392); Sabin-1 (AY184219), Sabin-2 (AY184220), Sabin-3 (AY184221); coxsackie virus 1 (AF499635), coxsackie virus 11 (AF499636), coxsackie virus 13 (AF465511), coxsackie virus 17 (AF499639), coxsackie virus 19 (AF499641), coxsackie virus 20 (AF499642, EF015012, and EF015019), coxsackie virus 21 (AF465515 and D00538), coxsackie virus 22 (AF499643), coxsackie virus 24 (EF026081); enterovirus C96 (HQ415759), enterovirus C99 (EF555644), enterovirus C102 (EF555645), enterovirus C104 (JX982259), enterovirus C105 (KM880098), enterovirus C109 (GQ865517), enterovirus C117 (JX262382), enterovirus C116 (JX514942), and enterovirus C118 (JX961709). cVDPV2, type 2 circulating vaccine-derived poliovirus; UTR, untranslated region; WPV, wild-type poliovirus.
1These authors contributed equally to this article.